ideal hosts should be:
- multiple cloning sites, T vector site style, selection marker
- capable of rapid growth in inexpensive medium
- nonpathogenic (virulence genes deleted)
- capable of incorporating DNA (able produce competent cells)
- gentically stable in culture (recA deleted, why? bc recA catalyzes homologous recombination, so you dont want that to happen since cloning DNA then put in ecoli plasmid, so nothing wild happens in ecoli)
- equipped with appropriate enzymes to allow replication of the vector
ideal hosts: escherichia coli (gram - bacteria, common cloning, k12 strain, can make electrorecian or other cells), Bacillus subtilis (gram +, easily to transform, doesnt need to make more cells), Saccharomyces cerevisiae (baker yeast)
write out? gives me a sequence of mamalian- clone this gene to a certain vector, expressed for ecoli
ex. gene for insulin, cloning need to do pcr,
/////
human insulin gene driven by ___
human promoter
so if put in ecoli -> ecoli cannot recognize human promoter, so need to put human promoter in there
ecoli cannot recognize exon or intro
so when cloning
need to do pcr
coding sequence as mRNA so ecoli ____
can recognize the introns and exons
start
...
Genetic engineering:
using in vitro techniques to alter genetic material in the laboratory
Basic techniques include:
- Restriction enzymes
- Gel electrophoresis
- Nucleic acid hybridization
- Molecular cloning
- Cloning vectors
Restriction enzymes:
recognize specific DNA sequences and cut DNA
Widespread among prokaryotes. Rare in eukaryotes
- Protect prokaryotes from hostile foreign DNA (e.g., viral genomes)
- Essential for in vitro DNA manipulation
Three classes of restriction enzymes:
Type II cleave DNA within their recognition sequence and are most useful for specific DNA manipulation
Restriction enzymes recognize ___
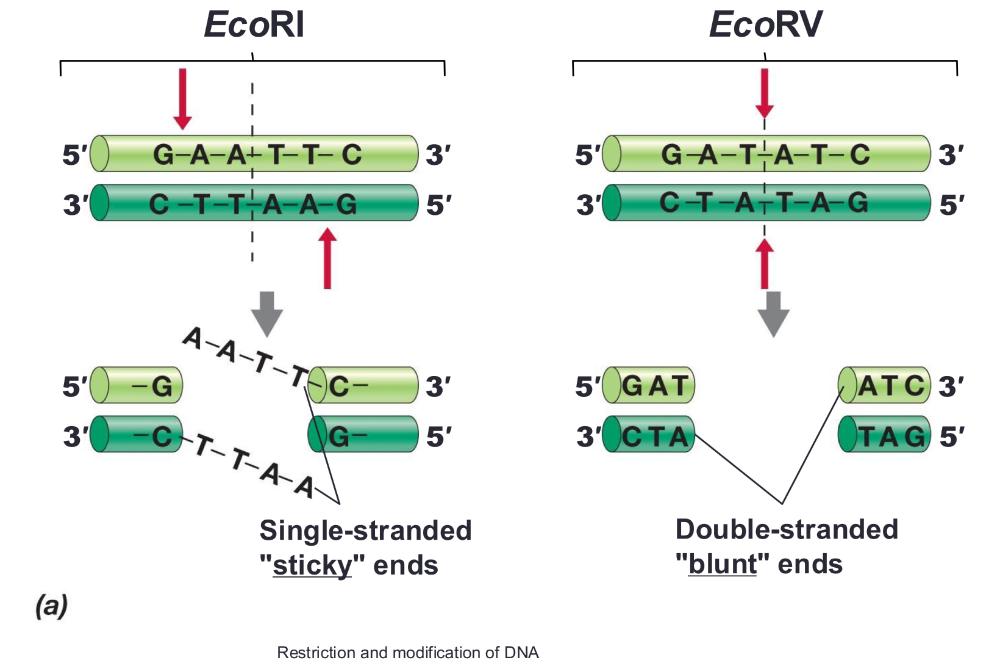
palindromes (inverted repeat sequences)
- Typically 4–8 base pairs long; EcoRI recognizes a 6-base-pair sequence
Sticky ends (allows mixing and matching) or blunt ends
Restriction enzymes protect cell from ___
invasion by foreign DNA
- Destroy foreign DNA
- Must protect their own DNA from inadvertent destruction
Modification enzymes:
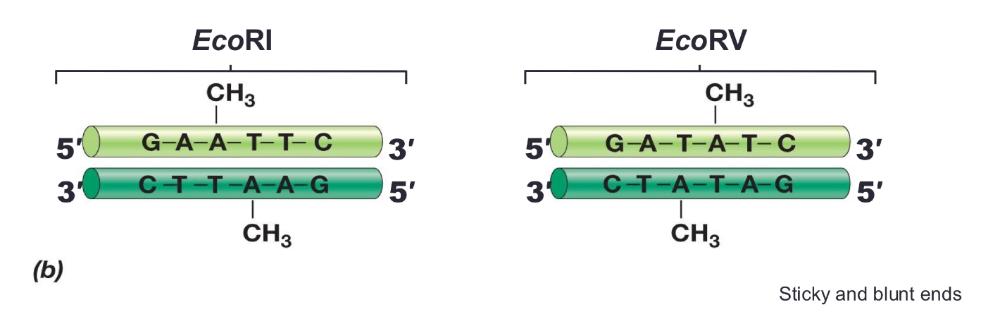
protect cell's DNA for restriction enzymes. Each restriction enzyme has a partner modification enzyme
- Chemically modify nucleotides in restriction recognition sequence
- Modification generally consists of methylation of DNA
Gel electrophoresis
separates DNA molecules based on size
- Electrophoresis uses an electrical field to separate charged molecules
- Gels are usually made of agarose, a polysaccharide
- Nucleic acids migrate through gel toward the positive electrode because of their negatively charged phosphate groups
Gels can be stained with ___

ethidium bromide (an intercalating agent) [slips int othe DNA bases], and DNA can be visualized under UV light
The same DNA that has been cut with different restriction enzymes will have
different banding patterns on an agarose gel
Size of fragments can be determined by comparison to a standard called a
DNA ladder
Nucleic acid hybridization
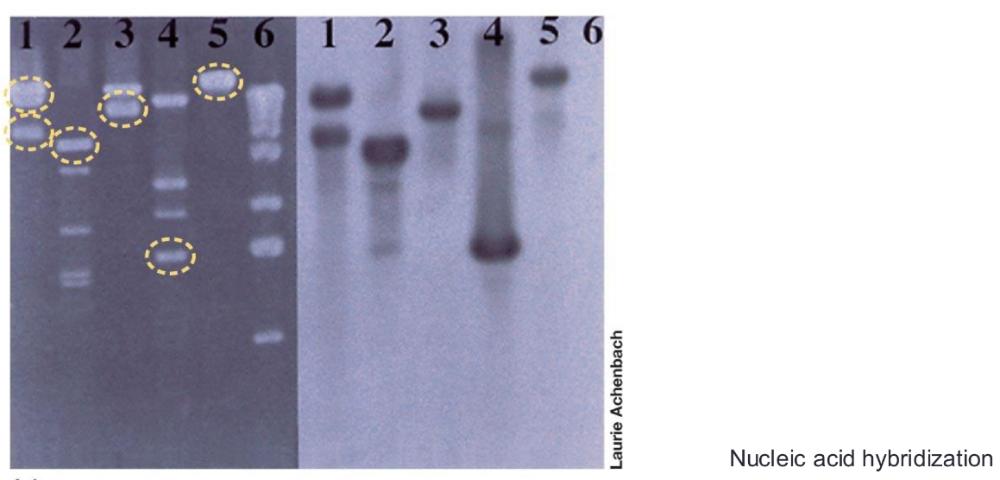
base pairing of single strands of DNA or RNA from two different sources to give a hybrid double helix
- Segment of single-stranded DNA that is used in hybridization and has a predetermined identity is called a nucleic acid probe
Southern blot
DNA as detecting targets
Northern blot
RNA as detecting targets
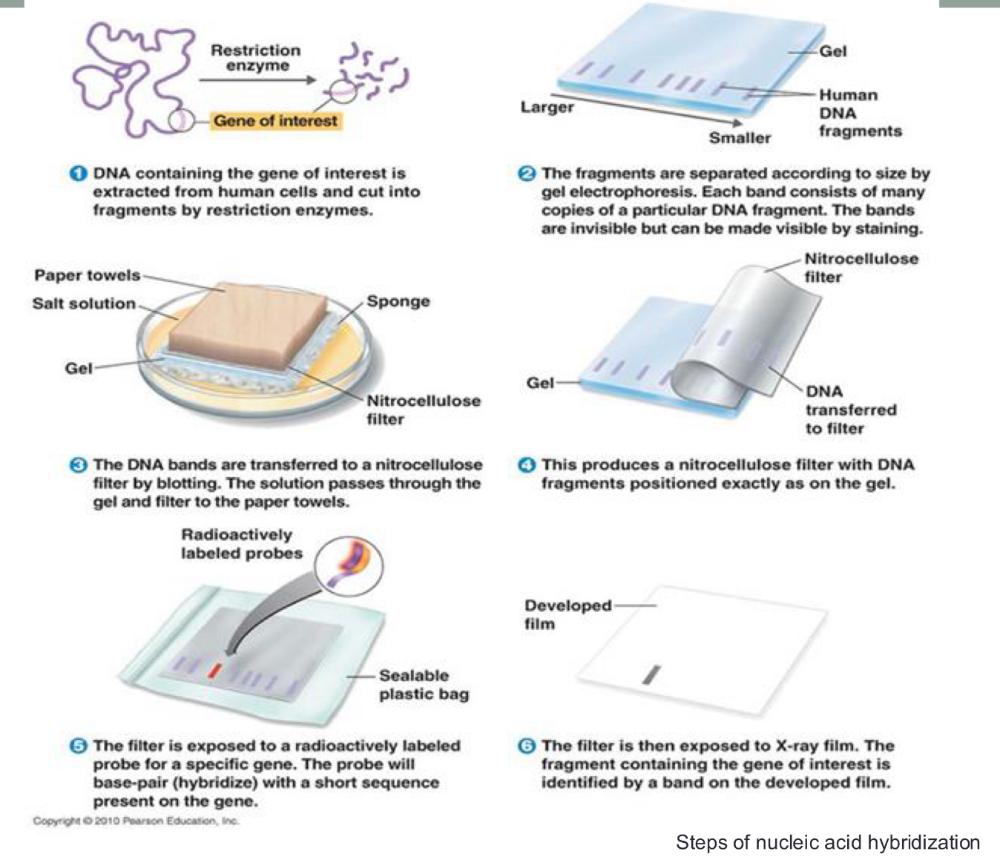
Steps of nucleic acid hybridization (photo)
FISH
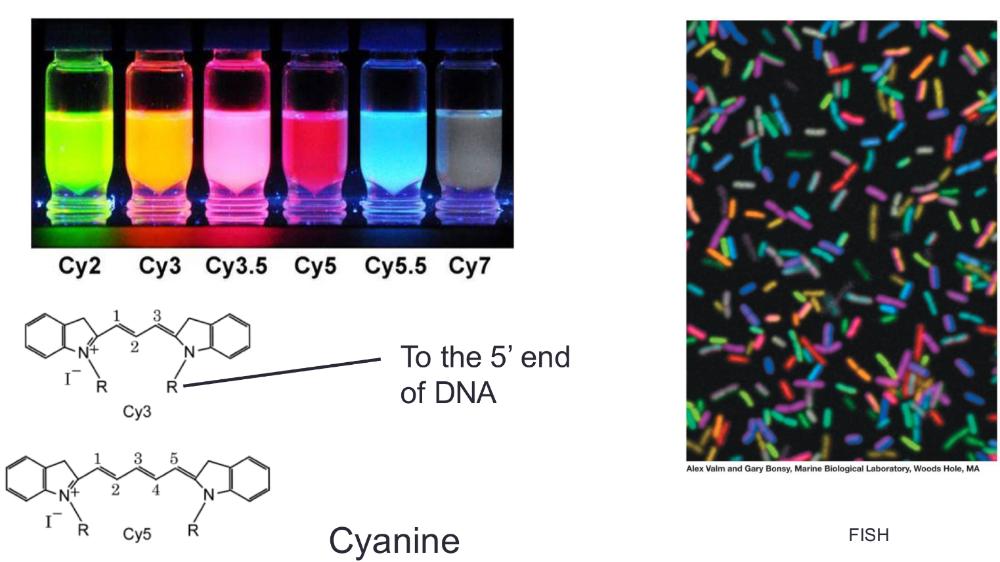
Fluorescent In Situ Hybridization (Figure 12.5)
- Uses fluorescent probe attached to oligonucleotide
polymerase chain reaction (PCR) amplification
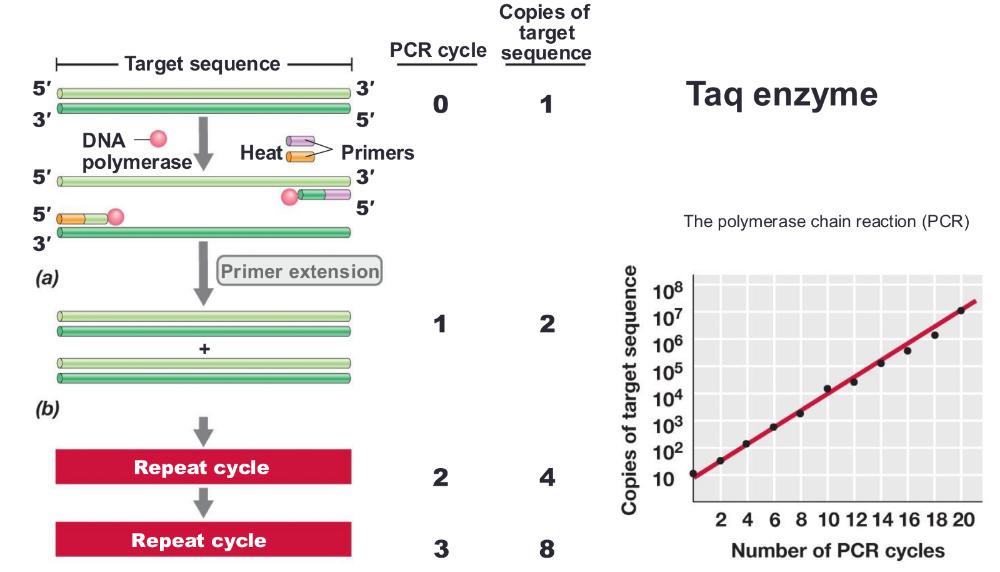
you can only amplify one strand
process:
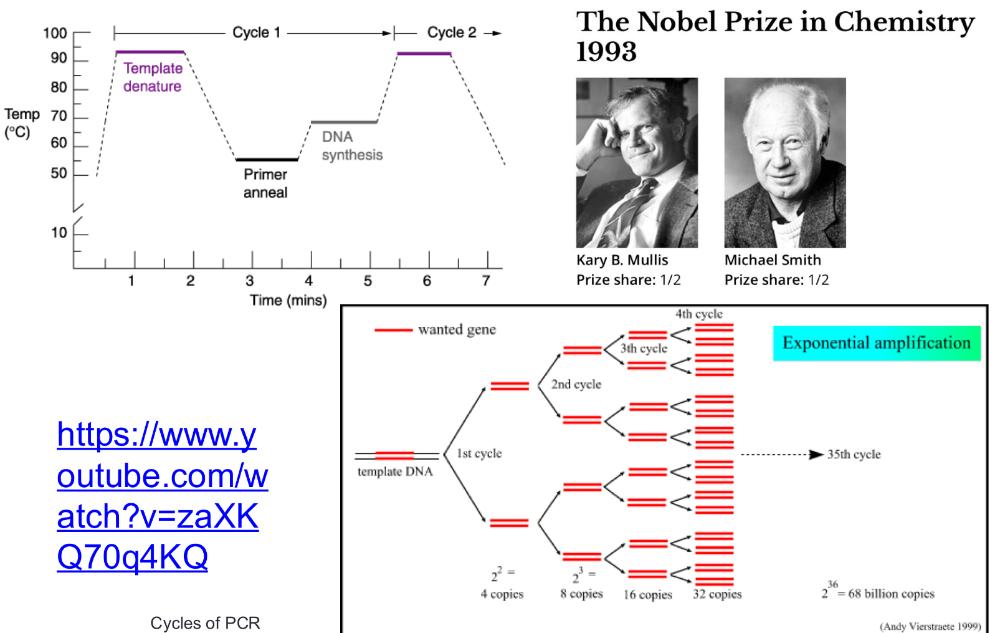
1. heat the DNA strand to melt it, separating strands, then cool so short DNA primers can bind to the target sequence (short DNA primers bind (or "anneal") to a single-stranded DNA template at a specific temperature= anneal primers)
2. then taq polymerase extend those primers, copying DNA
3. repeating cycle, anneal primers extend 20-30 times = exponential amplification
PCR cycles (photo)
How to design primers? (photo)
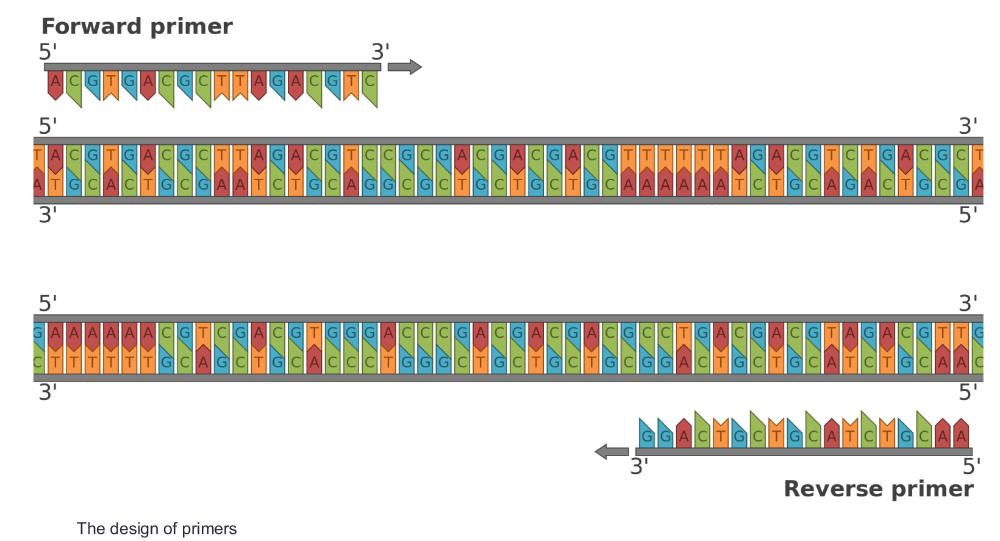
Primers are short DNA sequences that bind to specific regions on the template DNA to initiate replication—the forward primer binds to the bottom strand and runs 5'→3', while the reverse primer binds to the top strand in reverse complement, also running 5'→3'. This image shows how primers flank the target region, with the forward primer initiating synthesis to the right and the reverse primer to the left.

what are the primers?
start codon: ATG
stop codon: TAA
forward primer: first 18-20 bases of the coding strand
forward primer: ATGGACCAGTTCGATCAGA
reverse primer: TGCGGCGGAACTGCCG
Applications of PCR:
- Phylogenetic studies
- Surveying different groups of environmental organisms • Amplifying small amounts of DNA
- Identifying a specific bacteria
- Looking for a specific gene
Variations of PCR: (reverse transcription PCR)
Reverse transcription PCR (RT PCR)
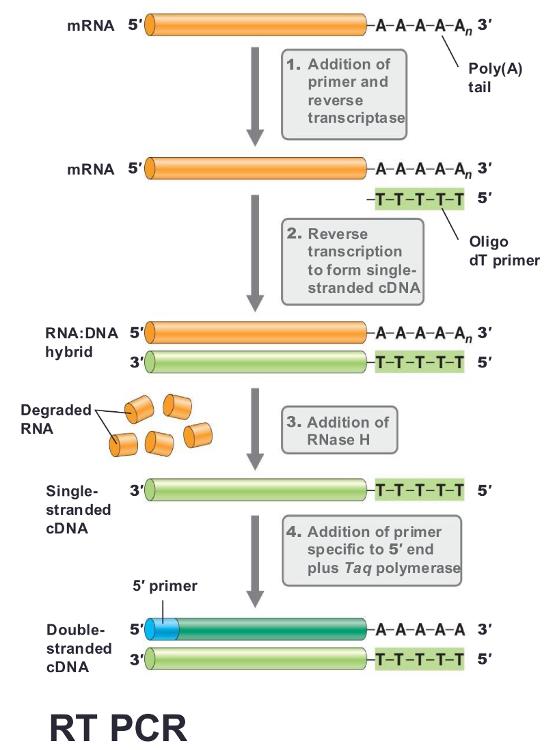
- Can make (c)DNA from an RNA template
- Uses the enzyme reverse transcriptase
often uses primer called oligo dt, binding in the poly-A tail found in most eukaryotic mRNA, then run PCR
Variations of PCR: (reverse transcription PCR)
Quantitative PCR (q PCR)
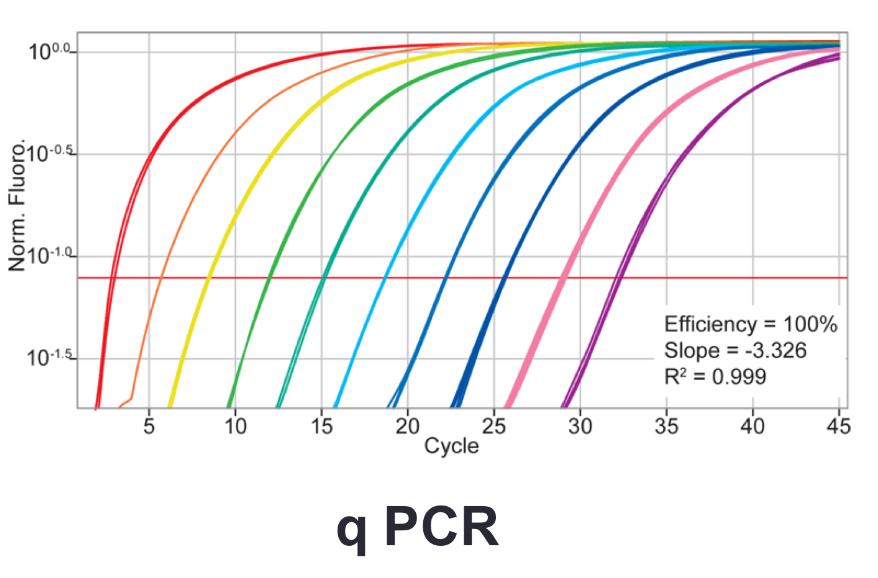
- Uses fluorescent probe to monitor the amplification process
how much of the target sequence was in the starting sample
Molecular cloning:
isolation and incorporation of a piece of DNA into a vector so it can be replicated and manipulated
Three main steps of gene cloning:
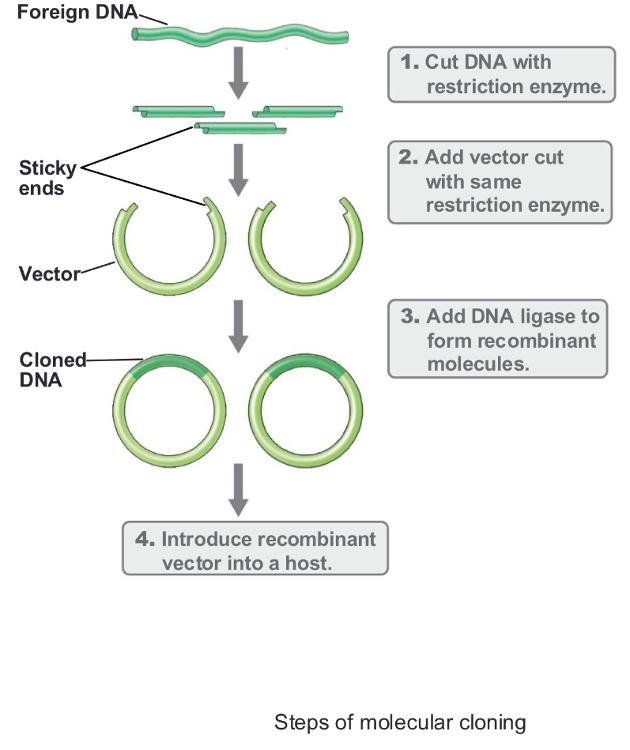
- Isolation and fragmentation of source DNA
- Insertion of DNA fragment into cloning vector (w sticky ends + DNA ligase)
- Introduction of (recombinant DNA) cloned DNA into host organism - usually in ecoli (transformation)
Steps of Molecular Cloning:
1. RCR/digestion:
Isolation and fragmentation of source DNA
- Source DNA can be genomic DNA, RNA, or PCR-amplified fragments
- Genomic DNA must first be restriction digested
Steps of Molecular Cloning:
2. Ligation:
Insertion of DNA fragment into cloning vector
- Most vectors are derived from plasmids or viruses
- DNA is generally inserted in vitro
- DNA ligase: enzyme that joins two DNA molecules
- Works with sticky or blunt ends
Steps of Molecular Cloning:
3. Transformation:
Introduction of cloned DNA into host organism
- Transformation is often used to get recombinant DNA into host
- Some cells will contain desired cloned gene, while other cells will have other cloned genes
• Gene library: mixture of cells containing a variety of genes
- Shotgun cloning: gene libraries made by cloning random genome fragments
Steps of Molecular Cloning:
4. Confirmation:
Detect the correct clones
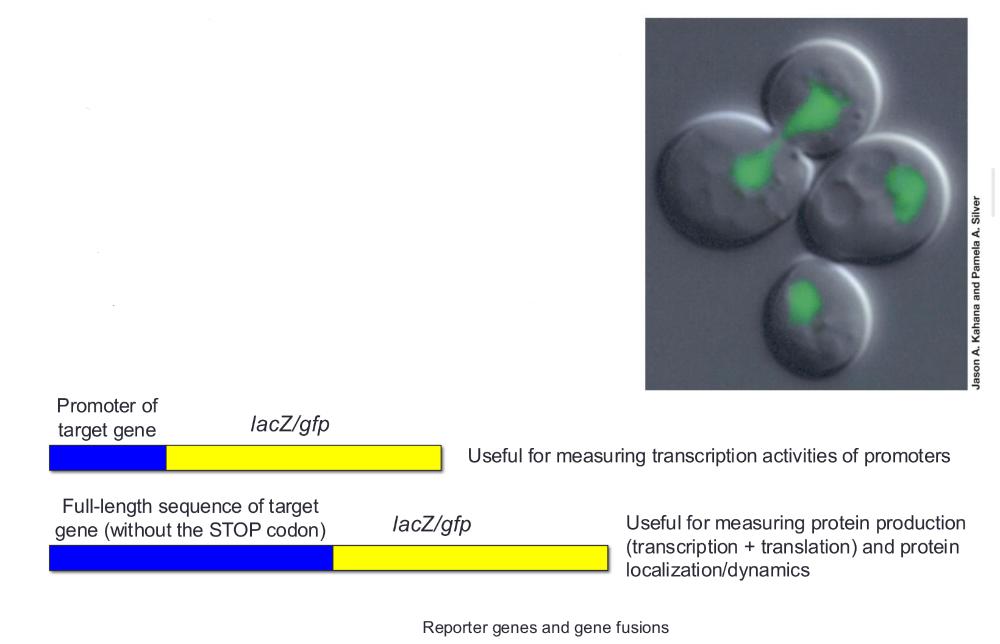
Reporter genes
Encode proteins that are easy to detect and assay
• Examples:
lacZ, luciferase, GFP genes
Gene fusions
Promoters or coding sequences of genes of interest can be swapped with those of reporter genes to elucidate gene regulation under various conditions
reporter genes and gene fusions (photo)
Plasmids are natural vectors and have useful properties as ___
cloning vectors
- Small size; easy to isolate DNA
- Independent origin of replication
- Multiple copy number; get multiple copies of cloned gene per cell
- Presence of selectable markers (usually antibiotic resistance)
Vector transfer is carried out by ___
chemical transformation or electroporation
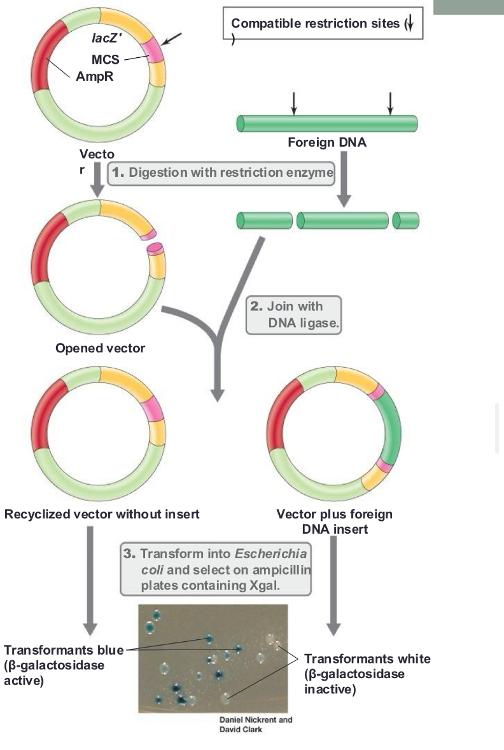
pUC18/19
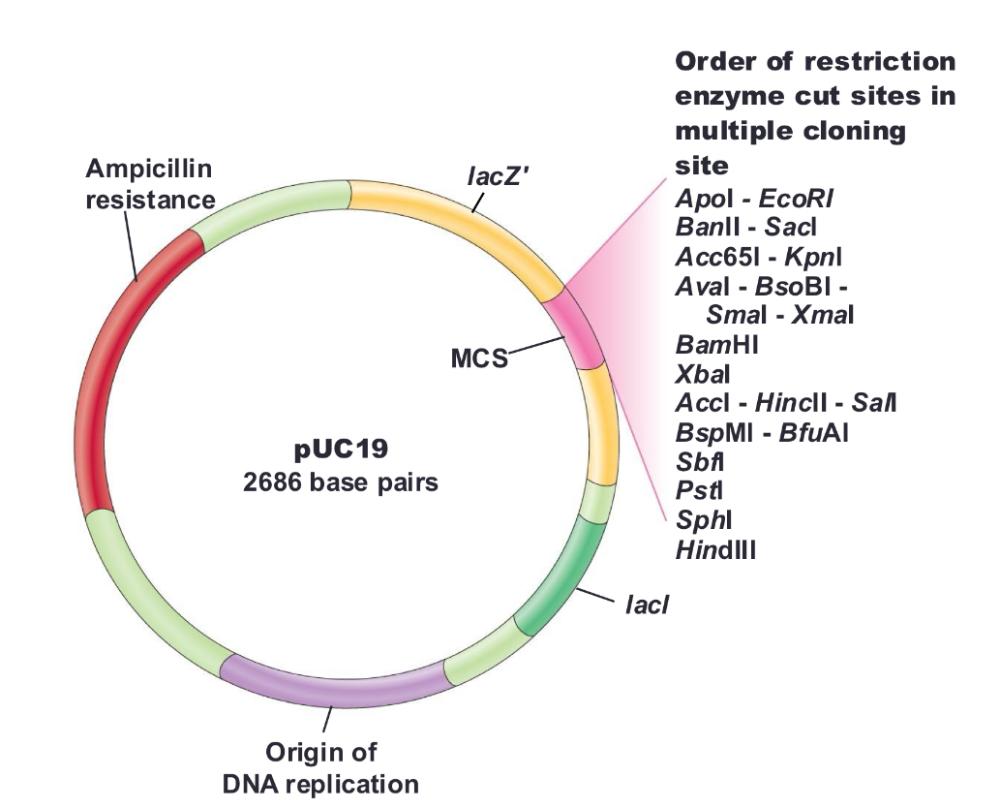
- Contains ampicillin-resistance and lacZ genes
- Contains multiple cloning site (MCS) within lacZ gene
need IPTG (inducer) to make sure lacz gene is turned on in the first place
Blue colonies do not have ___
vector with foreign DNA inserted
White colonies have ____
foreign DNA inserted (cloned gene containing)
Insertional inactivation:
lacZ gene is inactivated by insertion of foreign DNA
- Inactivated lacZ cannot process Xgal; blue color does not develop (IPTG needed to relieve the inhibition on the lacZ promoter)
blue/white screening (photo)
T vectors
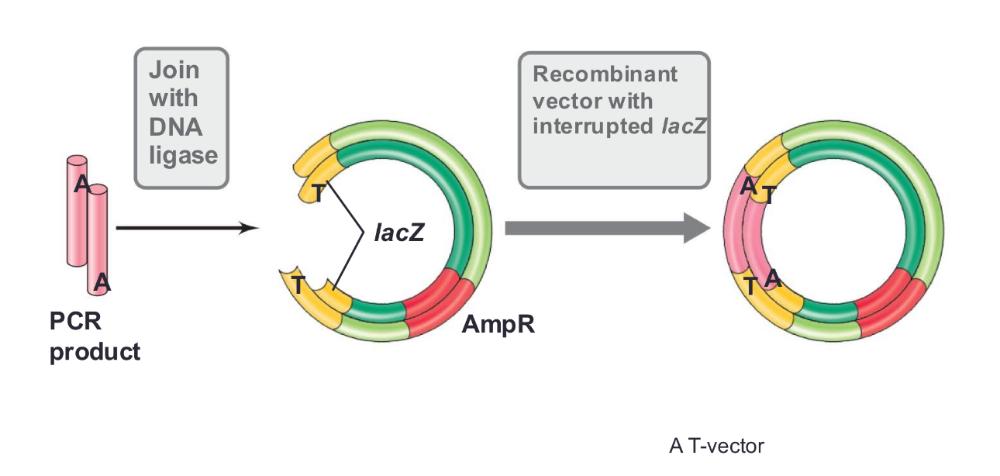
- Developed for cloning DNA products made by PCR (Figure 12.9)
- Taq adds an extra A at the 3’ ends of PCR products
Ideal hosts should be:
- Capable of rapid growth in inexpensive medium
- Nonpathogenic (virulence genes deleted)
- Capable of incorporating DNA (able produce competent cells)
- Genetically stable in culture (recA deleted, why? bc involved in recombination, creates deletions or insertions)
- Equipped with appropriate enzymes to allow replication of the vector
ex. Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae
Genotype of E. coli DH5α:
F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1
- F-: incapable of conjugation as donor
- lacZ: allows blue/white selection and β-galactosidase assay
- argF: blocks the production of arginine
- recA: lacks the recombinase
- endA: eliminates non-specific endonuclease activity, resulting in improved plasmid stability
- hsdR17: allows efficient transformation of DNA generated from PCR reactions *hsdR–eliminates restriction of unmethylated EcoKI sites.
- phoA: allows alkaline phosphotase assay
- supE: tRNA glutamine suppressor of amber (UAG)
- thi: requires thiamine for growth on minimal media
- gyrA: DNA gyrase mutant produces resistance to nalidixic acid
- relA: RNA is synthesized in absence of protein synthesis (relaxed phenotype) relA locus regulates the coupling between transcription and translation
Shuttle vectors:
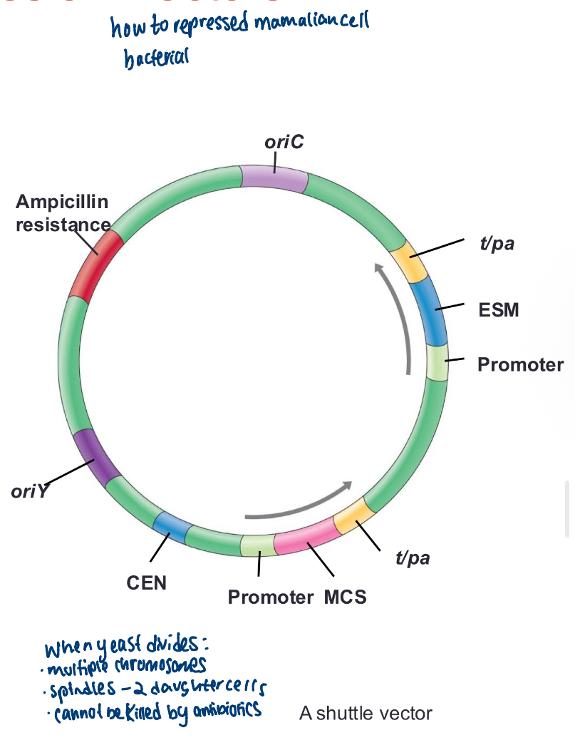
vectors that are stably maintained in two or more unrelated host organisms (e.g., E. coli and B. subtilis or E. coli and yeast)
• Bacterial plasmid engineered to function in eukaryotes
- Add a eukaryotic origin of replication
- Add a centromere recognition sequence(CEN)
- Add a eukaryotic selection marker (ESM)
- Add a eukaryotic RBS
- Optimize codon usage
- Eliminate introns using RT-PCR
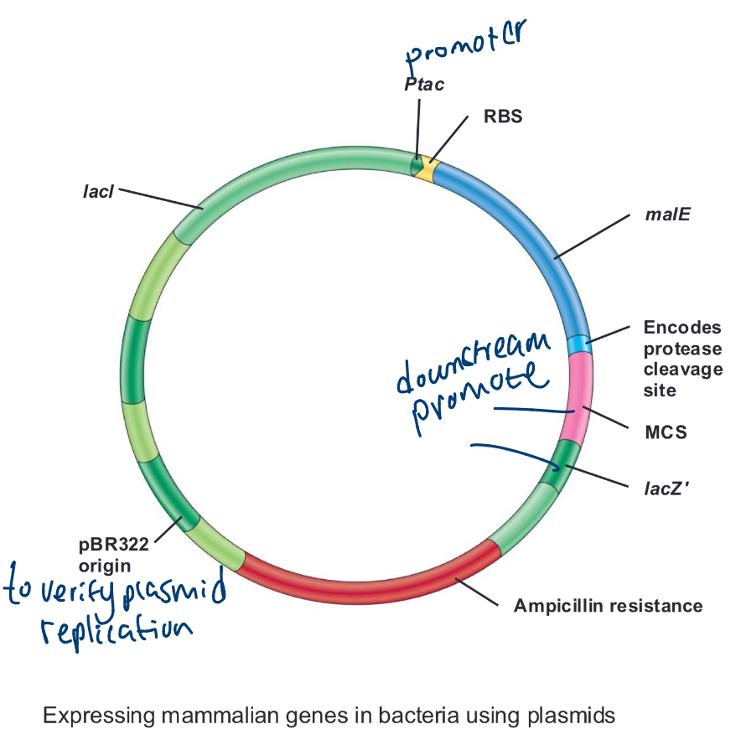
Expression vectors:
allow experimenter to control the expression of cloned genes (maximizing cloning production from gene)
- Based on transcriptional control
- Allow for high levels of protein expression
- Strong promoters
- Efficient operators (tight regulation)
- Effective transcription terminators are used to prevent expression of other genes on the plasmid
The T7 expression systems
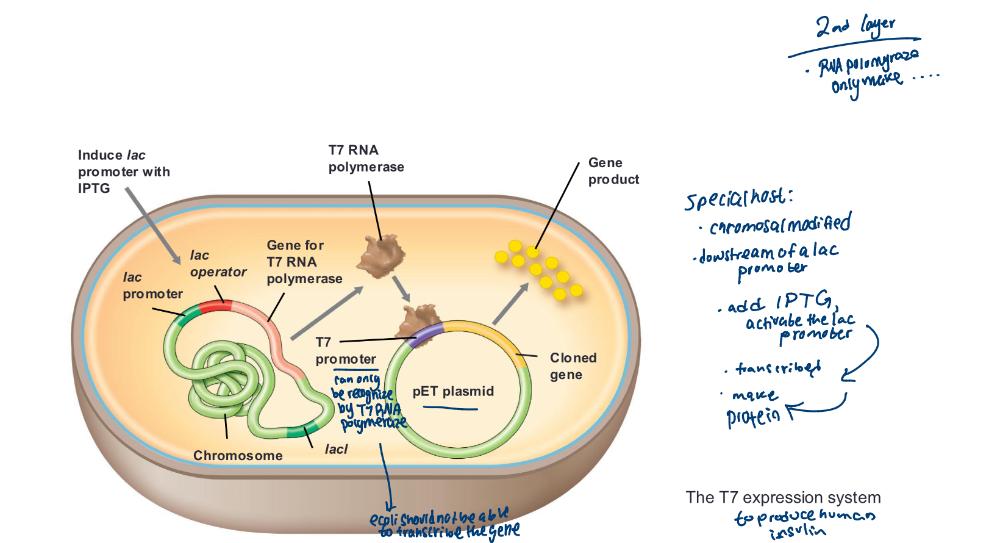
• In T7 expression vectors, cloned genes are placed under control of
the T7 promoter
• Gene for T7 RNA polymerase (recognized
by) present and under control of easily regulated system (e.g., lac)
- T7 RNA polymerase recognizes only T7 promoters
• Transcribes only cloned genes
• Shuts down host transcription
homework

homework 2
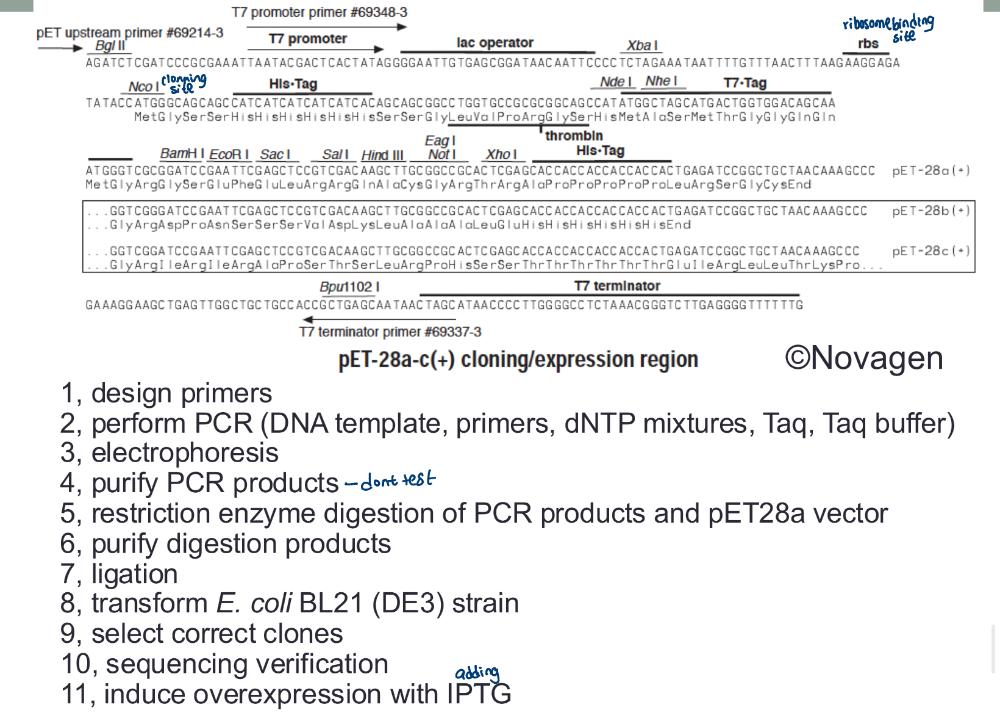
Expressing Mammalian Genes in Bacteria:
Biotechnology (plants)
Use of living organisms for industrial or commercial applications
Expressing Mammalian Genes in Bacteria:
Genetically modified organism (GMO)
Organism whose genome has been altered
Genetic engineering allows expression of _____
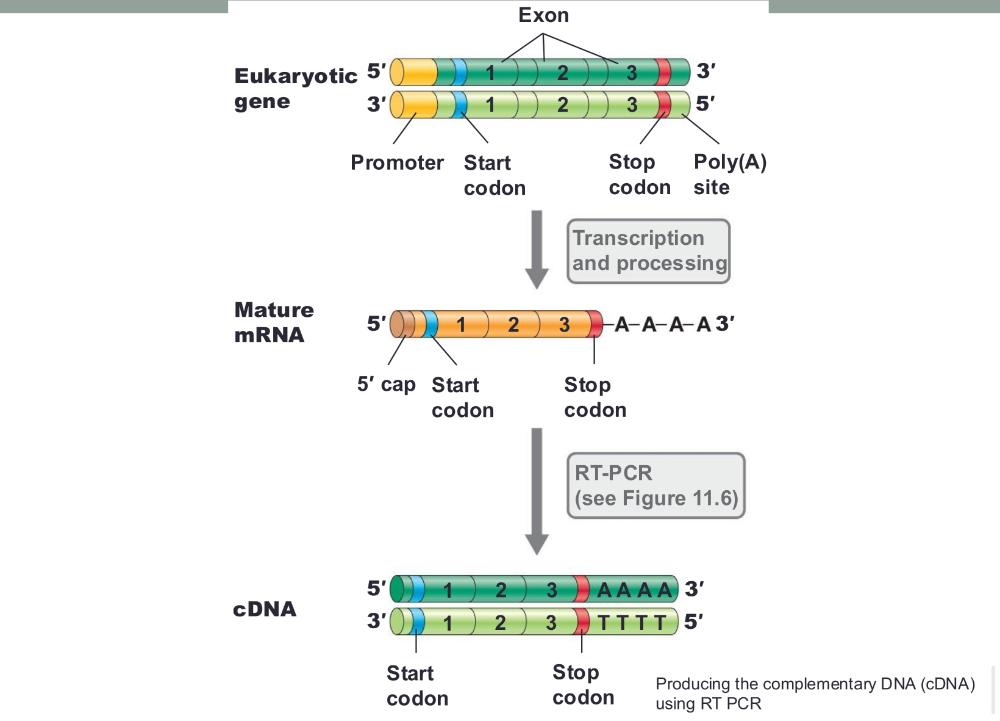
eukaryotic genes in prokaryotes (e.g., insulin)
• This is achieved by cloning the gene via mRNA
Producing the complementary DNA (cDNA) using RT PCR (photo)
Exp-ressing Mammalian Genes in Bacteria:
Protein synthesis in a foreign host is subject to _____
other problems
• Degradation by intracellular proteases
• Toxicity to prokaryotic host
• Formation of inclusion bodies (folds)
Exp-ressing Mammalian Genes in Bacteria:
Fusion of a target protein with a carrier protein facilitates ______
protein purification
Expressing mammalian genes in bacteria using plasmids (photo)
Insulin was the first human protein made commercially by ______
genetic engineering
Somatotropin
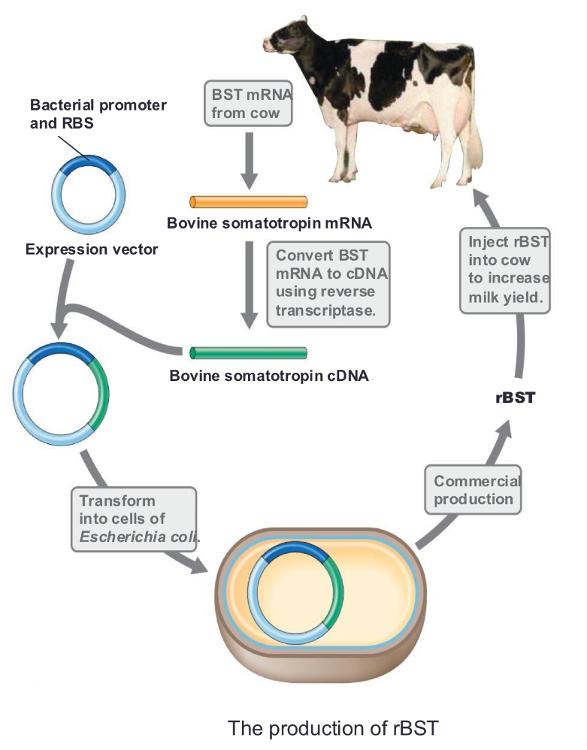
a growth hormone, is another widely produced hormone
- Cloned as cDNA from the mRNA
- Recombinant bovine
somatotropin (rBST) is commonly used in the dairy
industry; stimulates milk production in cows
rBST is a growth hormone for milk
Transgenic Organisms in Agriculture and Aquaculture:
Transgenic organism
Organism that contains a gene from another organism (can modify animal or plant, micro injection)
Plants can be genetically modified through several approaches, including:
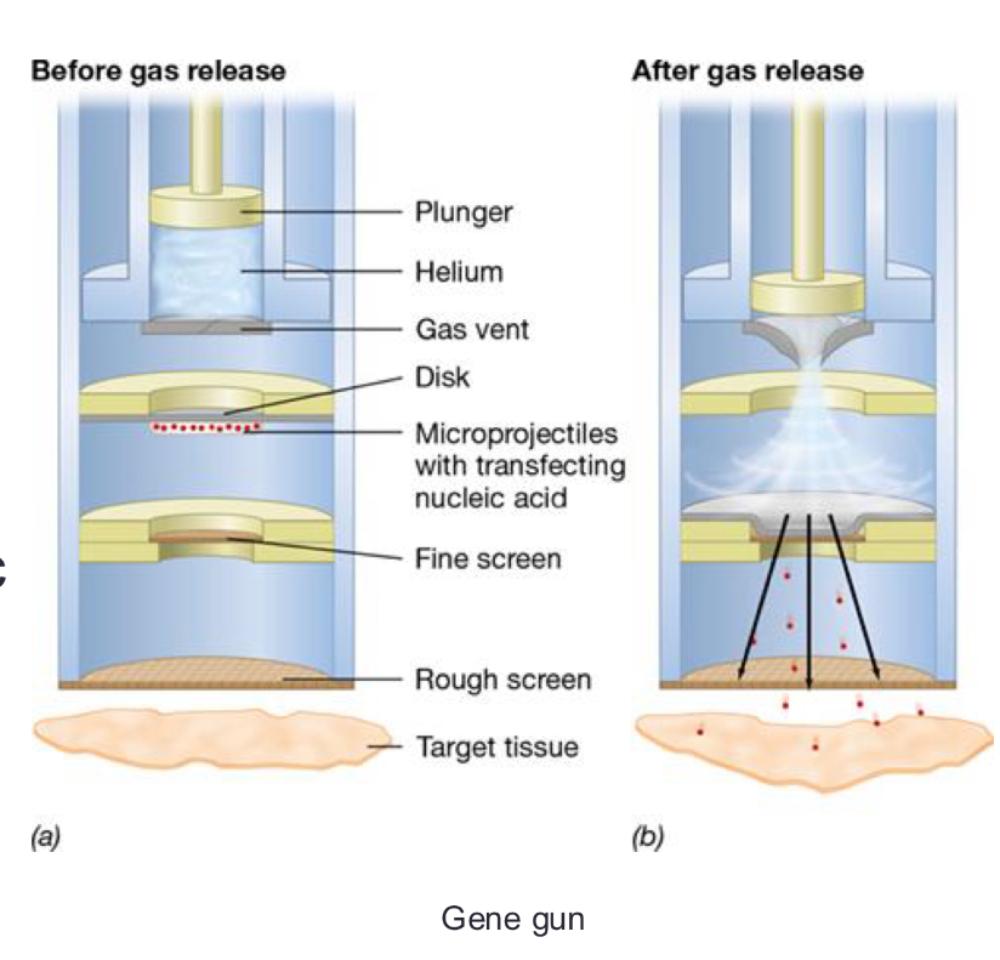
- Electroporation
- Particle gun methods (gene gun)
- Use of plasmids from bacterium Agrobacterium tumefaciens
Many successes in plant genetic engineering; several transgenic plants are in agricultural production
The plant pathogen Agrobacterium tumefaciens can be used to _____
introduce DNA into plants
A. tumefaciens contains the ____
Ti plasmid, which is responsible for virulence
The Ti plasmid contains genes that ____
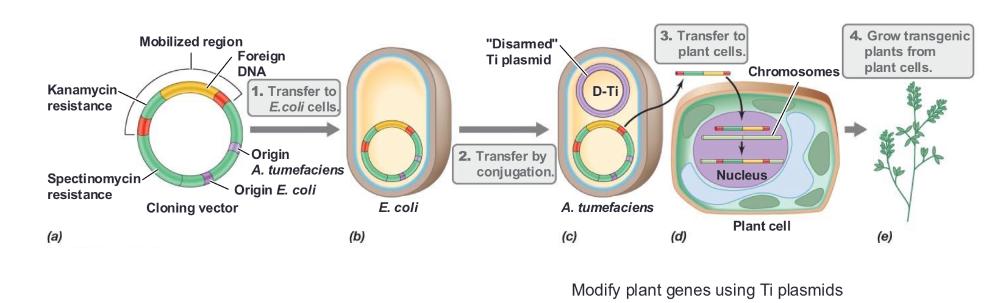
mobilize DNA for transfer to the plant
The segment of the Ti plasmid that is transferred to the plant is called the _____
T-DNA
Several areas are targeted for genetic improvements in plants, including _____
resistance to herbicides, insects, and microbial disease, as well as improved product quality
Plants are engineered to have ______ to protect them from herbicides applied to kill weeds (e.g., glyphosate)
herbicide resistance
One widely used approach for genetically engineering ______ in plants involves introducing genes encoding the toxic protein of Bacillus thuringiensis (Bt toxin)
insect resistance
Transgenic animals are useful for improving ___
livestock and other animals for human consumption
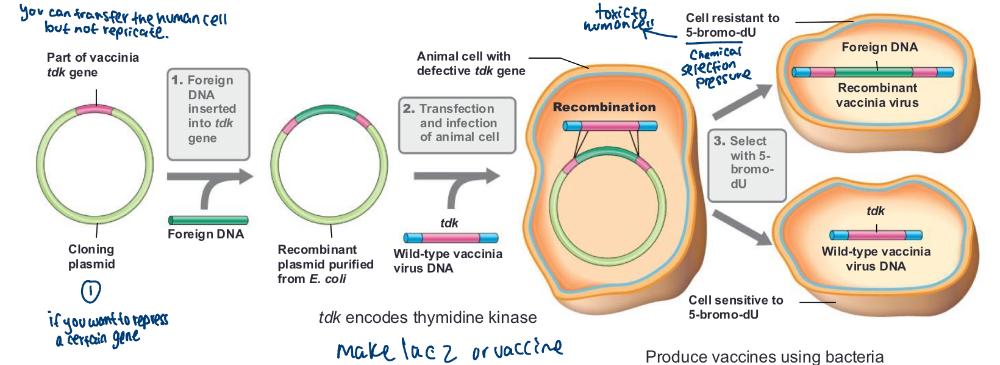
Recombinant vaccines are _____
Vector vaccine and Subunit vaccine
Vector vaccine
vaccine made by inserting genes from a pathogenic virus into a relatively harmless carrier virus (e.g., vaccinia virus)
Subunit vaccine
contain only a specific protein or proteins from a pathogenic organism
Polyvalent vaccine
A single vaccine that immunizes against two different diseases
produce vaccines using bacteria (photo)
Gene mining
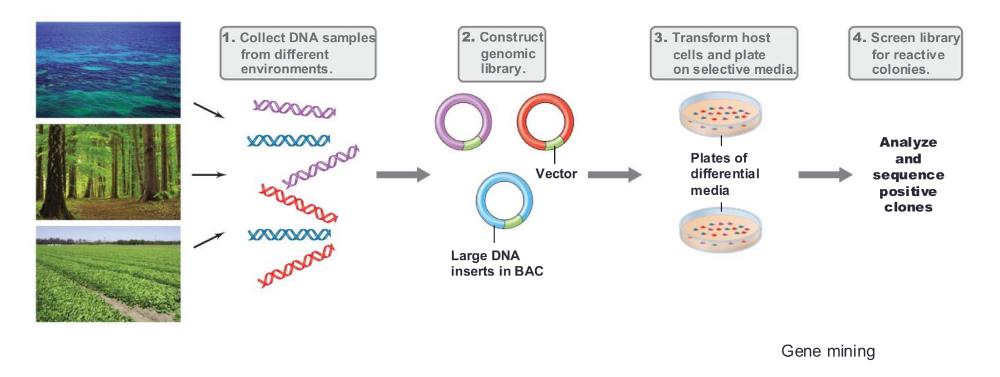
- The process of isolating potentially useful novel genes from the environment without culturing the organism
- To do so, DNA (or RNA) is directly isolated from the environment and cloned into appropriate expression vectors BACs (bacterial artificial chromosomes), and the library is screened for activities of interest
- pathway engineering
gene mining drawing
Engineering Metabolic Pathways
The production of small metabolites by genetic engineering typically involves multiple genes that must be expressed in a coordinated manner
Pathway engineering
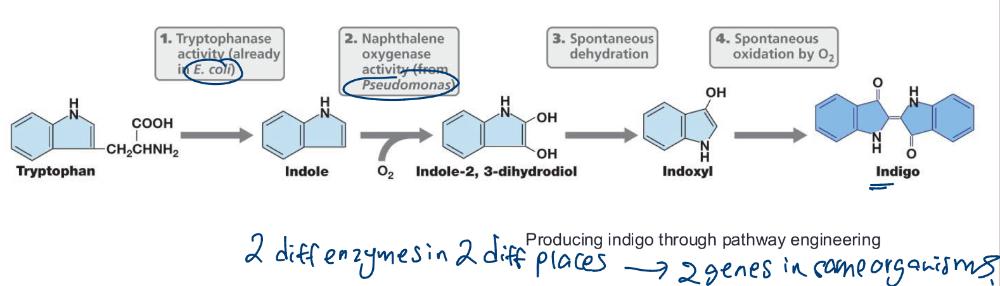
The process of assembling a new or improved biochemical pathway using genes from one or more organisms (e.g., indigo)
Synthetic biology
using genetic engineering to create novel
biological
systems out of available parts (biobricks)
Examples
- Self-replicating synthetic bacterium
- Genetically modified E. coli that produces photographs
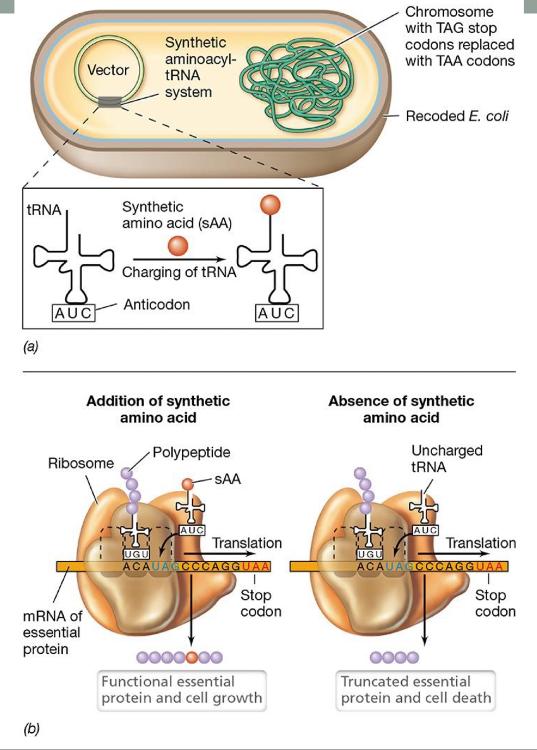
Biocontainment of Genetically Modified Organisms (GMO genome)
- recode so bacterium can only grow if supplied with a synthetic amino acid
- replaced all TAG stop codons with TAA
- express genes for an aminoacyl-tRNA synthetase that recognizes synthetic amino acid (sAA) and a tRNA with an AUC anticodon
- essential genes were modified to contain TAG codon but not affect activity
Recoding and control of genetically modified Escherichia coli