Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
lec 16-18 biotechnology and synthetic biology
front 1 ideal hosts should be: | back 1
ideal hosts: escherichia coli (gram - bacteria, common cloning, k12 strain, can make electrorecian or other cells), Bacillus subtilis (gram +, easily to transform, doesnt need to make more cells), Saccharomyces cerevisiae (baker yeast) |
front 2 write out? gives me a sequence of mamalian- clone this gene to a certain vector, expressed for ecoli ex. gene for insulin, cloning need to do pcr, | back 2 ///// |
front 3 human insulin gene driven by ___ | back 3 human promoter so if put in ecoli -> ecoli cannot recognize human promoter, so need to put human promoter in there ecoli cannot recognize exon or intro |
front 4 so when cloning | back 4 need to do pcr |
front 5 coding sequence as mRNA so ecoli ____ | back 5 can recognize the introns and exons |
front 6 start | back 6 no data |
front 7 Genetic engineering: | back 7 using in vitro techniques to alter genetic material in the laboratory Basic techniques include:
|
front 8 Restriction enzymes: | back 8 recognize specific DNA sequences and cut DNA Widespread among prokaryotes. Rare in eukaryotes
|
front 9 Three classes of restriction enzymes: | back 9 Type II cleave DNA within their recognition sequence and are most useful for specific DNA manipulation |
front 10 Restriction enzymes recognize ___ | back 10 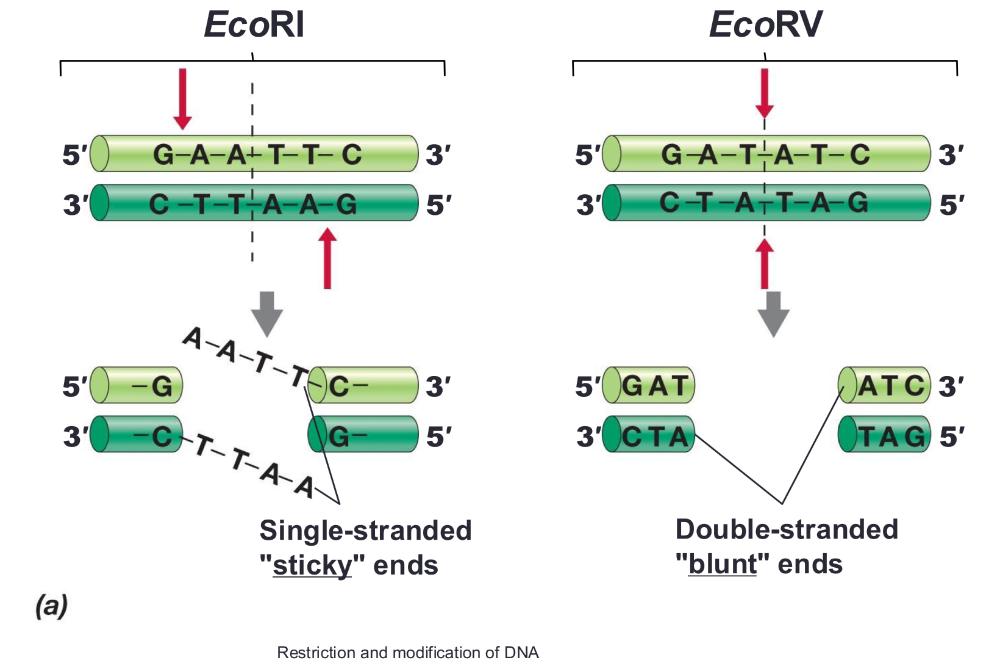 palindromes (inverted repeat sequences)
Sticky ends (allows mixing and matching) or blunt ends |
front 11 Restriction enzymes protect cell from ___ | back 11 invasion by foreign DNA
|
front 12 Modification enzymes: | back 12 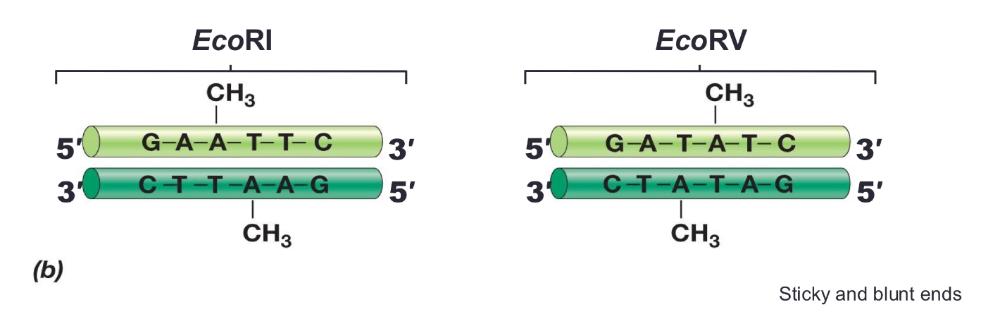 protect cell's DNA for restriction enzymes. Each restriction enzyme has a partner modification enzyme
|
front 13 Gel electrophoresis | back 13 separates DNA molecules based on size
|
front 14 Gels can be stained with ___ | back 14  ethidium bromide (an intercalating agent) [slips int othe DNA bases], and DNA can be visualized under UV light |
front 15 The same DNA that has been cut with different restriction enzymes will have | back 15 different banding patterns on an agarose gel |
front 16 Size of fragments can be determined by comparison to a standard called a | back 16  DNA ladder |
front 17 Nucleic acid hybridization | back 17 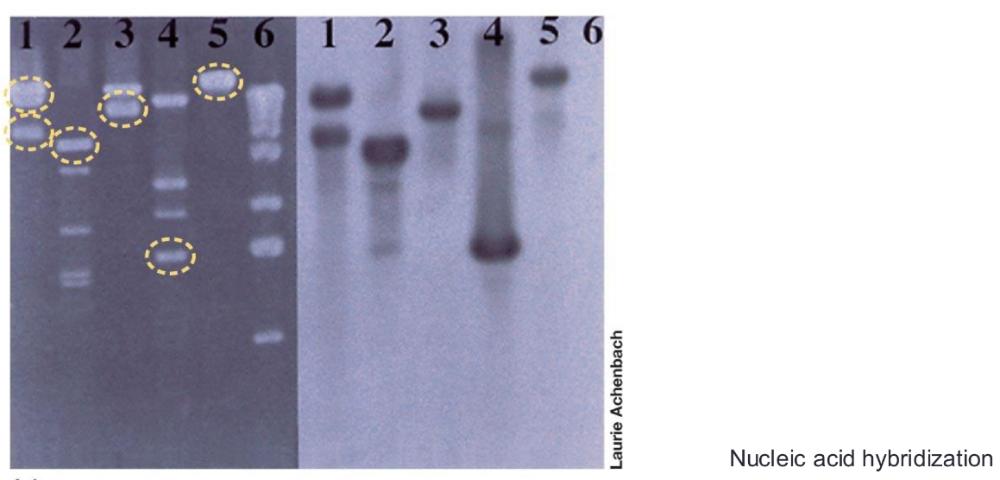 base pairing of single strands of DNA or RNA from two different sources to give a hybrid double helix
|
front 18 Southern blot | back 18 DNA as detecting targets |
front 19 Northern blot | back 19 RNA as detecting targets |
front 20 Steps of nucleic acid hybridization (photo) | back 20 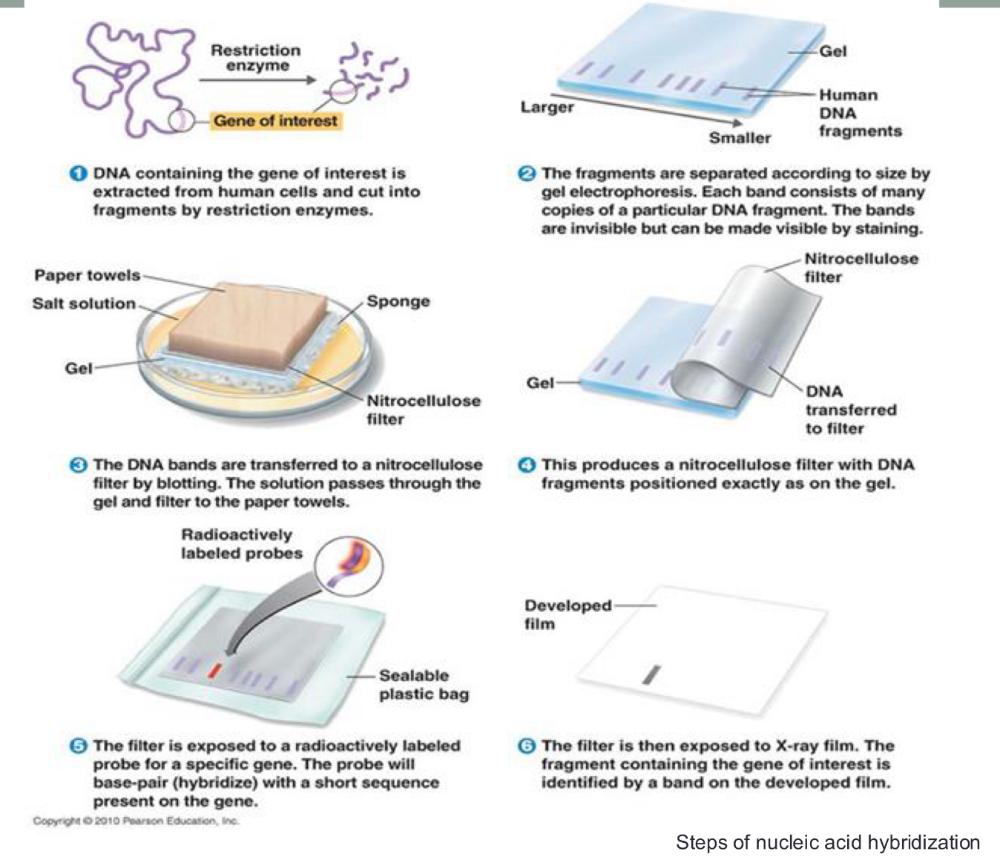 |
front 21 FISH | back 21 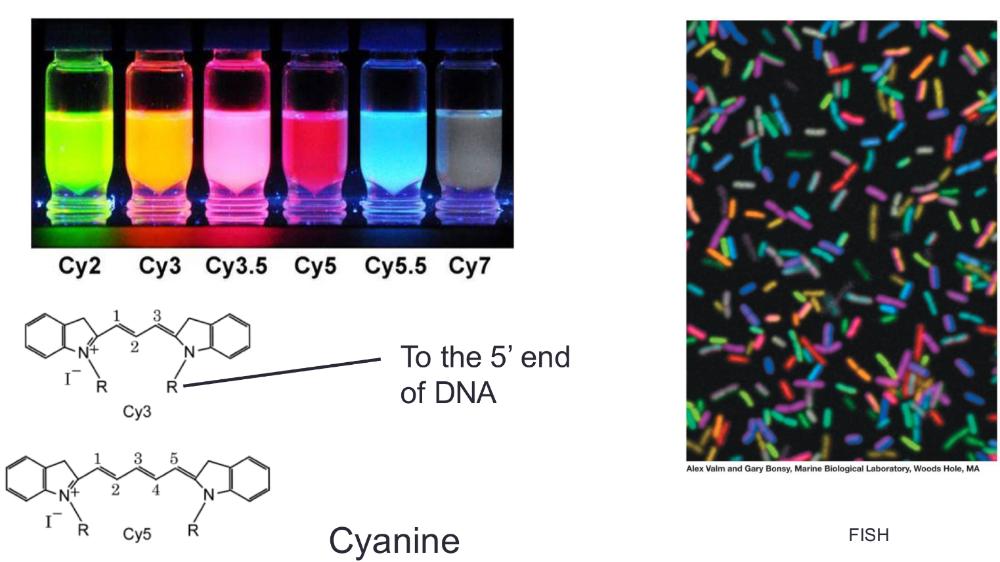 Fluorescent In Situ Hybridization (Figure 12.5)
|
front 22 polymerase chain reaction (PCR) amplification | back 22 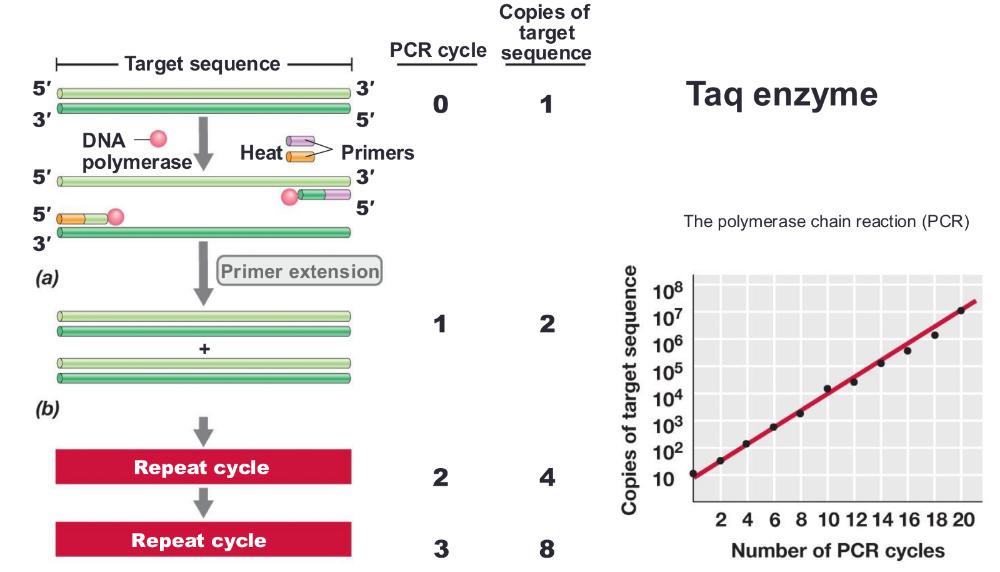 you can only amplify one strand process: 1. heat the DNA strand to melt it, separating strands, then cool so short DNA primers can bind to the target sequence (short DNA primers bind (or "anneal") to a single-stranded DNA template at a specific temperature= anneal primers) 2. then taq polymerase extend those primers, copying DNA 3. repeating cycle, anneal primers extend 20-30 times = exponential amplification |
front 23 PCR cycles (photo) | back 23 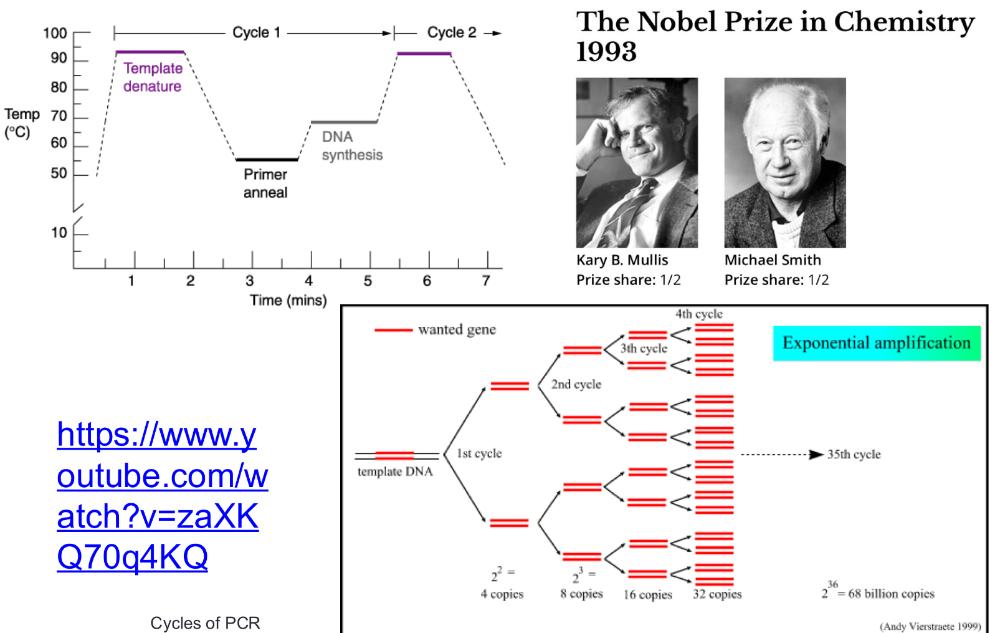 |
front 24 How to design primers? (photo) | back 24 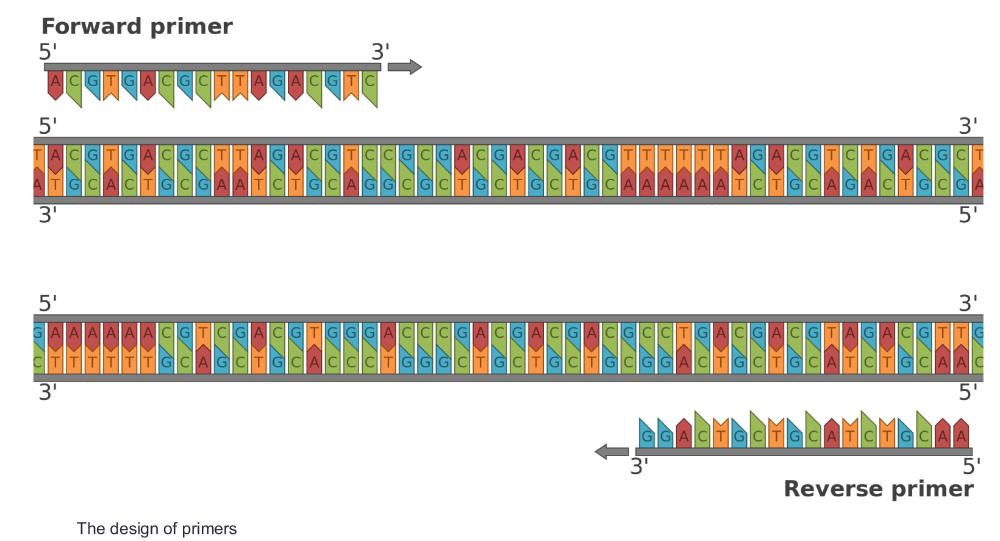 Primers are short DNA sequences that bind to specific regions on the template DNA to initiate replication—the forward primer binds to the bottom strand and runs 5'→3', while the reverse primer binds to the top strand in reverse complement, also running 5'→3'. This image shows how primers flank the target region, with the forward primer initiating synthesis to the right and the reverse primer to the left. |
front 25  what are the primers? | back 25 start codon: ATG stop codon: TAA forward primer: first 18-20 bases of the coding strand forward primer: ATGGACCAGTTCGATCAGA reverse primer: TGCGGCGGAACTGCCG |
front 26 Applications of PCR: | back 26
|
front 27 Variations of PCR: (reverse transcription PCR) Reverse transcription PCR (RT PCR) | back 27 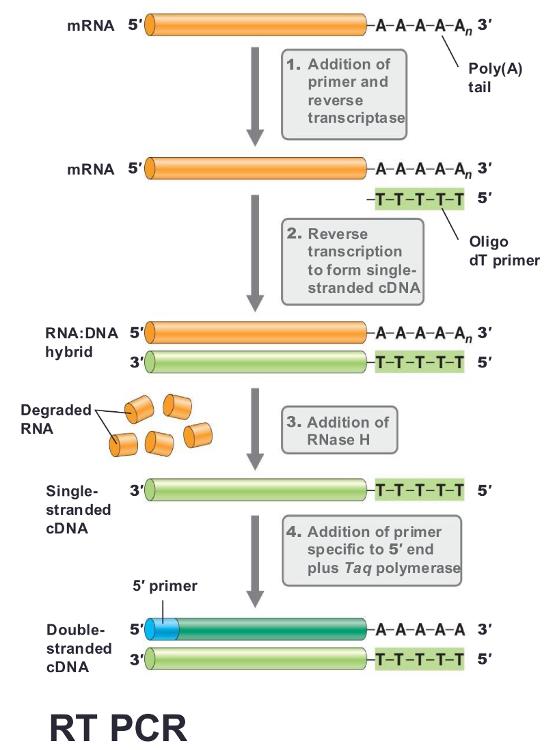
often uses primer called oligo dt, binding in the poly-A tail found in most eukaryotic mRNA, then run PCR |
front 28 Variations of PCR: (reverse transcription PCR) Quantitative PCR (q PCR) | back 28 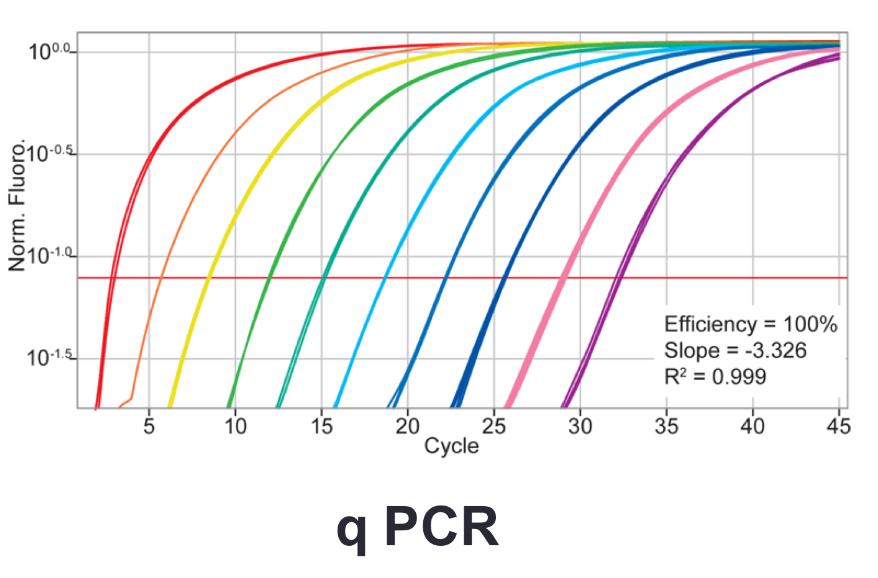
how much of the target sequence was in the starting sample |
front 29 Molecular cloning: | back 29 isolation and incorporation of a piece of DNA into a vector so it can be replicated and manipulated |
front 30 Three main steps of gene cloning: | back 30 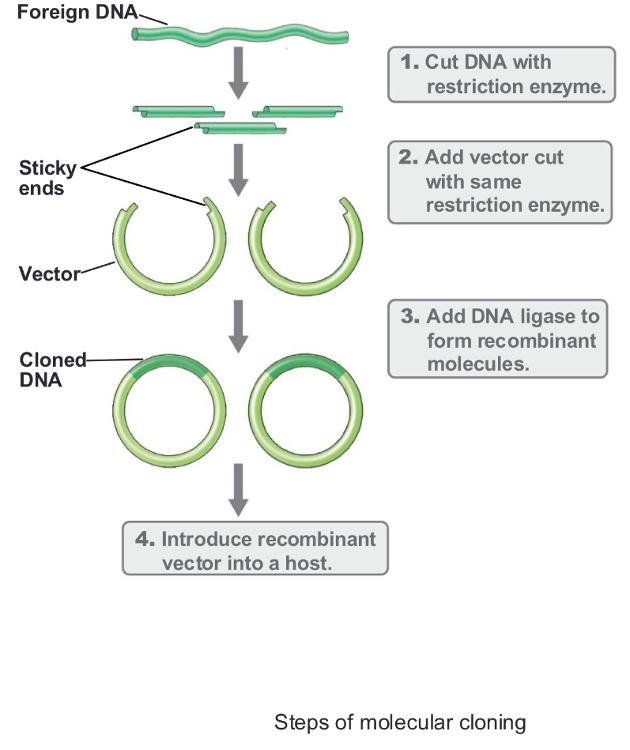
|
front 31 Steps of Molecular Cloning: 1. RCR/digestion: | back 31 Isolation and fragmentation of source DNA
|
front 32 Steps of Molecular Cloning: 2. Ligation: | back 32 Insertion of DNA fragment into cloning vector
- Works with sticky or blunt ends |
front 33 Steps of Molecular Cloning: 3. Transformation: | back 33 Introduction of cloned DNA into host organism
• Gene library: mixture of cells containing a variety of genes
|
front 34 Steps of Molecular Cloning: 4. Confirmation: | back 34 Detect the correct clones |
front 35 Reporter genes | back 35 Encode proteins that are easy to detect and assay |
front 36 Gene fusions | back 36 Promoters or coding sequences of genes of interest can be swapped with those of reporter genes to elucidate gene regulation under various conditions |
front 37 reporter genes and gene fusions (photo) | back 37 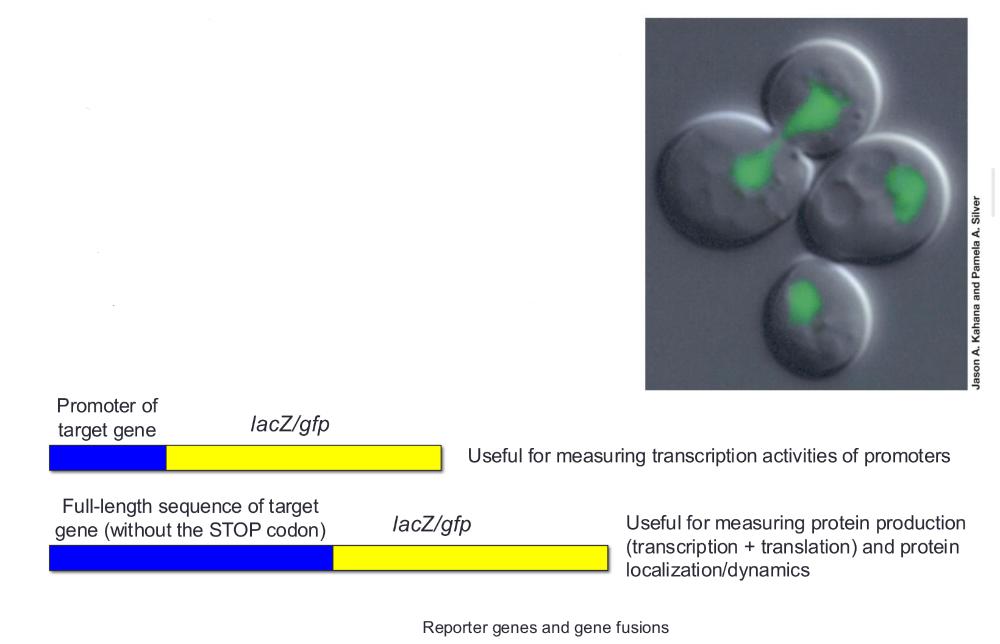 |
front 38 Plasmids are natural vectors and have useful properties as ___ | back 38 cloning vectors
|
front 39 Vector transfer is carried out by ___ | back 39 chemical transformation or electroporation |
front 40 pUC18/19 | back 40 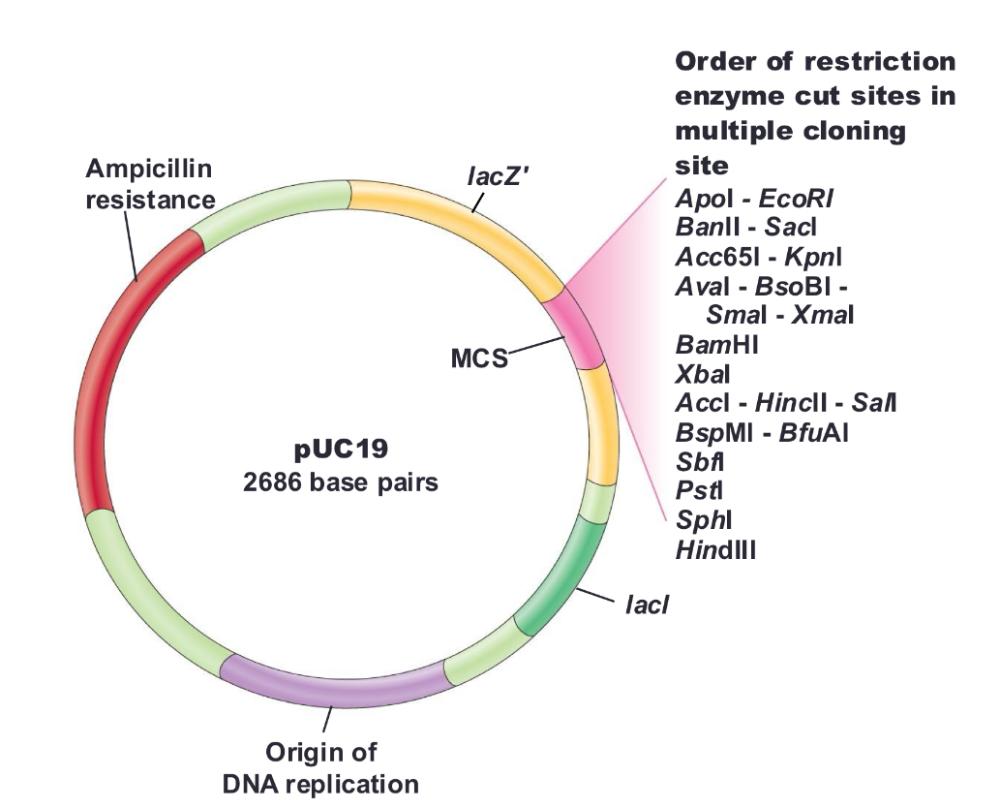
need IPTG (inducer) to make sure lacz gene is turned on in the first place |
front 41 Blue colonies do not have ___ | back 41 vector with foreign DNA inserted |
front 42 White colonies have ____ | back 42 foreign DNA inserted (cloned gene containing) |
front 43 Insertional inactivation: | back 43 lacZ gene is inactivated by insertion of foreign DNA
|
front 44 blue/white screening (photo) | back 44 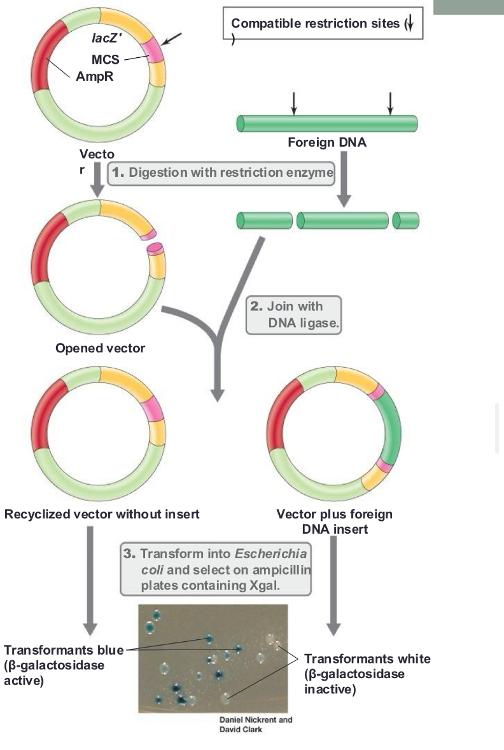 |
front 45 T vectors | back 45 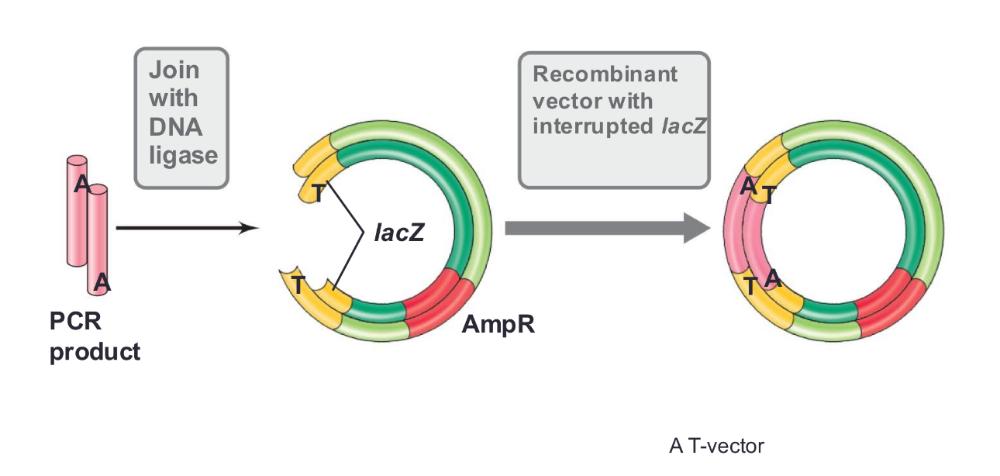
|
front 46 Ideal hosts should be: | back 46
ex. Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae |
front 47 Genotype of E. coli DH5α: | back 47 F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1
|
front 48 Shuttle vectors: | back 48 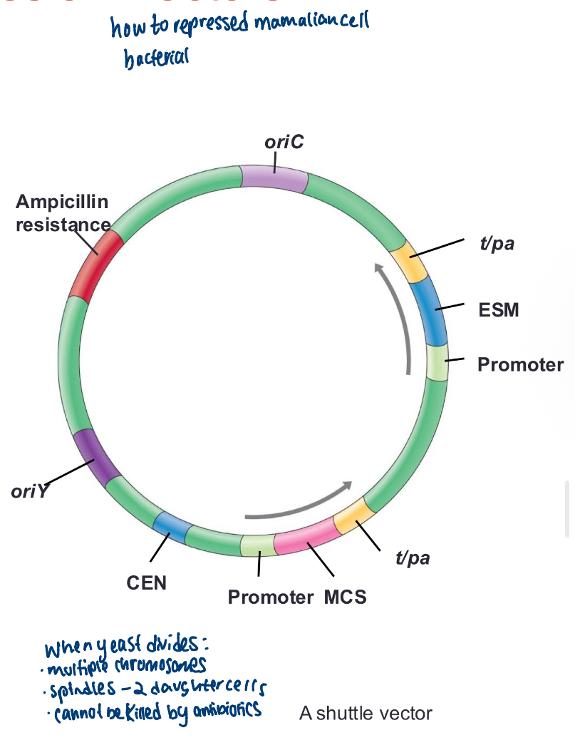 vectors that are stably maintained in two or more unrelated host organisms (e.g., E. coli and B. subtilis or E. coli and yeast) • Bacterial plasmid engineered to function in eukaryotes
|
front 49 Expression vectors: | back 49 allow experimenter to control the expression of cloned genes (maximizing cloning production from gene)
|
front 50 The T7 expression systems | back 50 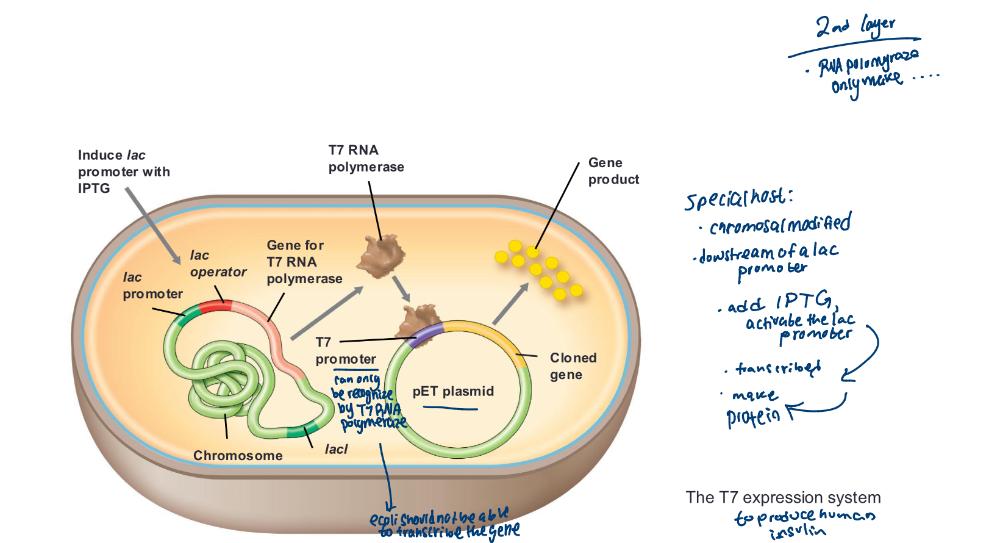 • In T7 expression vectors, cloned genes are placed under control of
the T7 promoter
|
front 51 homework | back 51  |
front 52 homework 2 | back 52 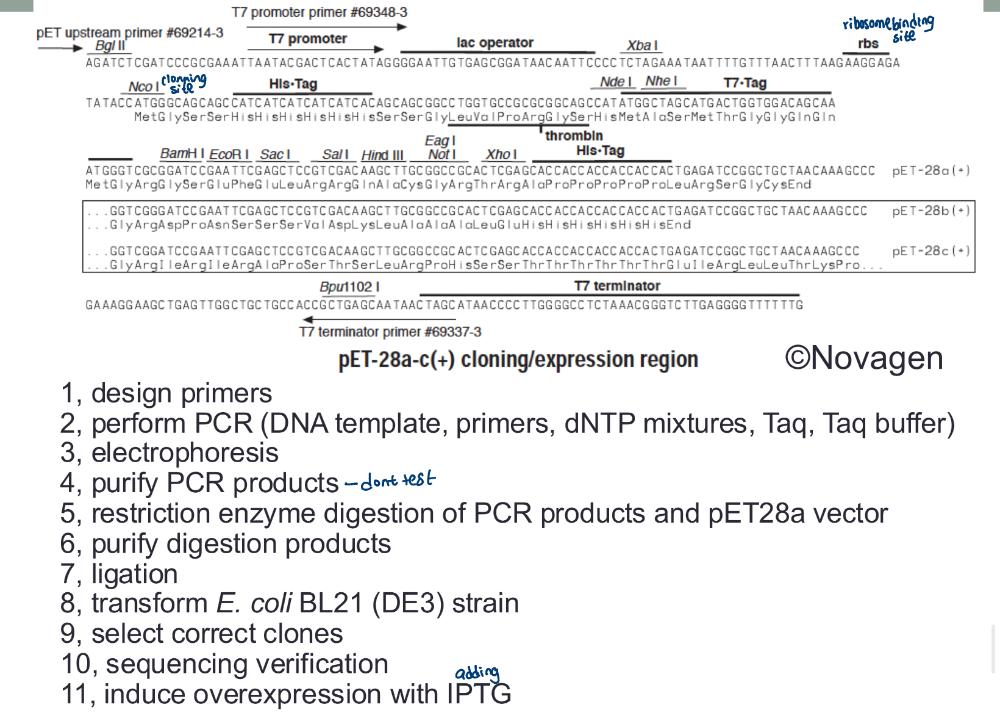 |
front 53 Expressing Mammalian Genes in Bacteria: Biotechnology (plants) | back 53 Use of living organisms for industrial or commercial applications |
front 54 Expressing Mammalian Genes in Bacteria: Genetically modified organism (GMO) | back 54 Organism whose genome has been altered |
front 55 Genetic engineering allows expression of _____ | back 55 eukaryotic genes in prokaryotes (e.g., insulin) • This is achieved by cloning the gene via mRNA |
front 56 Producing the complementary DNA (cDNA) using RT PCR (photo) | back 56 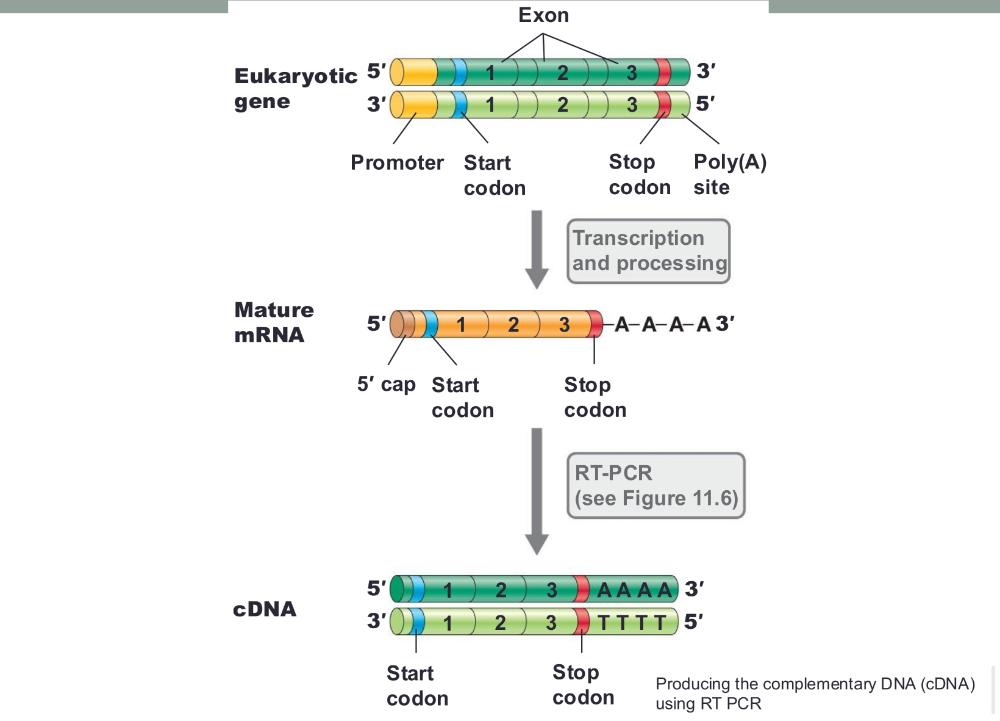 |
front 57 Exp-ressing Mammalian Genes in Bacteria: Protein synthesis in a foreign host is subject to _____ | back 57 other problems • Degradation by intracellular proteases • Toxicity to prokaryotic host |
front 58 Exp-ressing Mammalian Genes in Bacteria: Fusion of a target protein with a carrier protein facilitates ______ | back 58 protein purification |
front 59 Expressing mammalian genes in bacteria using plasmids (photo) | back 59 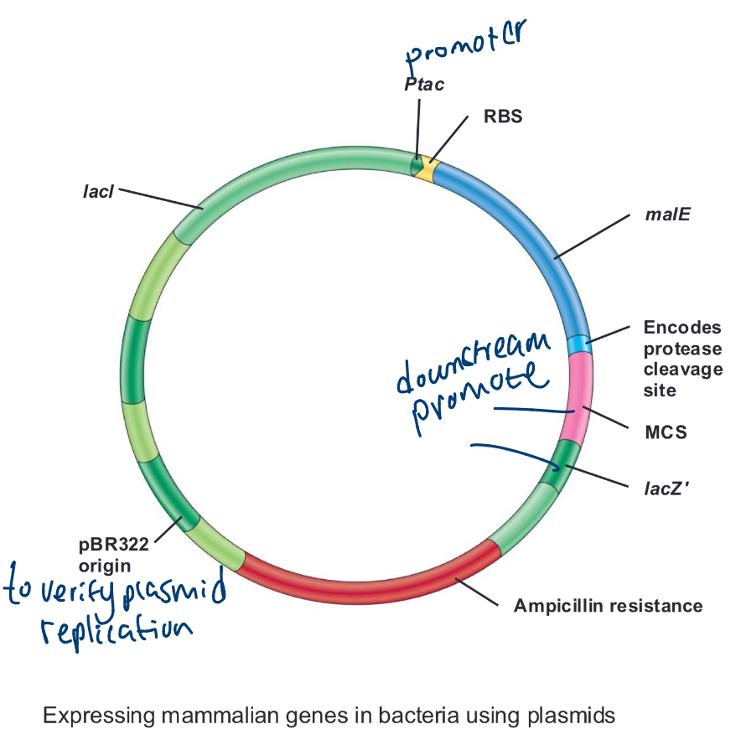 |
front 60 Insulin was the first human protein made commercially by ______ | back 60 genetic engineering |
front 61 Somatotropin | back 61 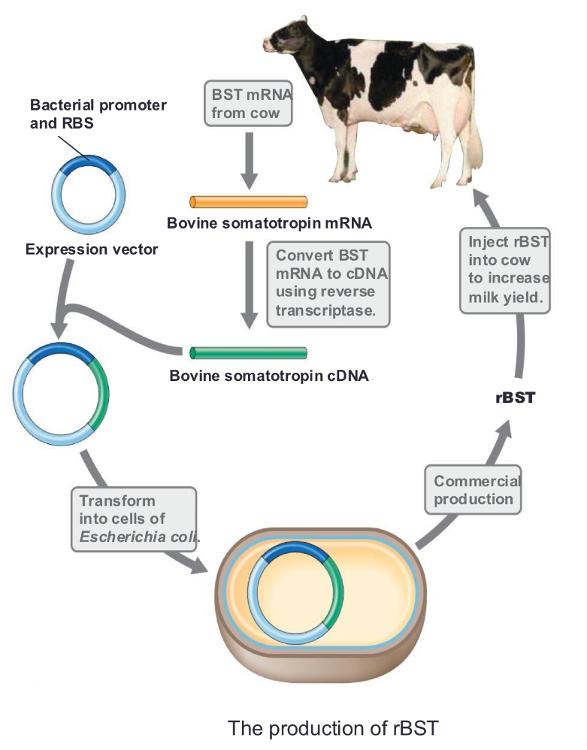 a growth hormone, is another widely produced hormone
rBST is a growth hormone for milk |
front 62 Transgenic Organisms in Agriculture and Aquaculture: Transgenic organism | back 62 Organism that contains a gene from another organism (can modify animal or plant, micro injection) |
front 63 Plants can be genetically modified through several approaches, including: | back 63 
Many successes in plant genetic engineering; several transgenic plants are in agricultural production |
front 64 The plant pathogen Agrobacterium tumefaciens can be used to _____ | back 64 introduce DNA into plants |
front 65 A. tumefaciens contains the ____ | back 65 Ti plasmid, which is responsible for virulence |
front 66 The Ti plasmid contains genes that ____ | back 66 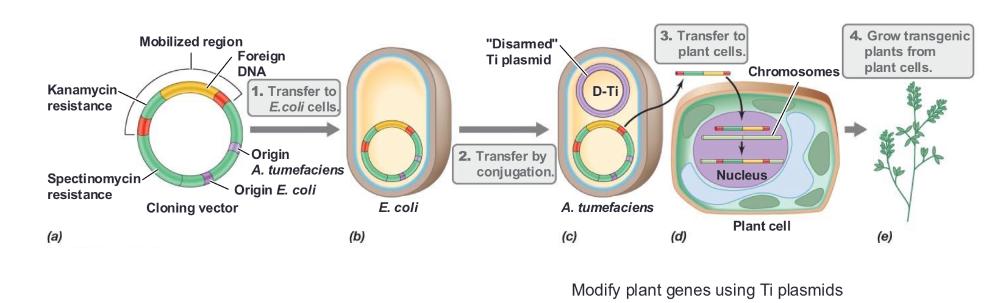 mobilize DNA for transfer to the plant |
front 67 The segment of the Ti plasmid that is transferred to the plant is called the _____ | back 67 T-DNA |
front 68 Several areas are targeted for genetic improvements in plants, including _____ | back 68 resistance to herbicides, insects, and microbial disease, as well as improved product quality |
front 69 Plants are engineered to have ______ to protect them from herbicides applied to kill weeds (e.g., glyphosate) | back 69 herbicide resistance |
front 70 One widely used approach for genetically engineering ______ in plants involves introducing genes encoding the toxic protein of Bacillus thuringiensis (Bt toxin) | back 70 insect resistance |
front 71 Transgenic animals are useful for improving ___ | back 71 livestock and other animals for human consumption |
front 72 Recombinant vaccines are _____ | back 72 Vector vaccine and Subunit vaccine |
front 73 Vector vaccine | back 73 vaccine made by inserting genes from a pathogenic virus into a relatively harmless carrier virus (e.g., vaccinia virus) |
front 74 Subunit vaccine | back 74 contain only a specific protein or proteins from a pathogenic organism |
front 75 Polyvalent vaccine | back 75 A single vaccine that immunizes against two different diseases |
front 76 produce vaccines using bacteria (photo) | back 76 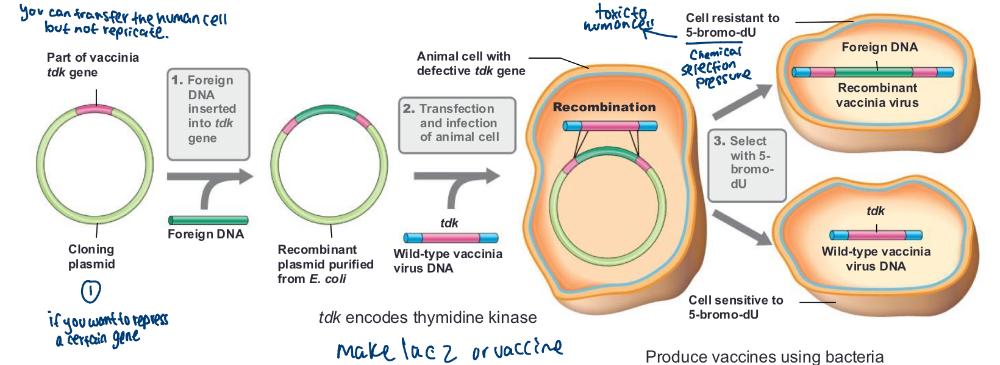 |
front 77 Gene mining | back 77 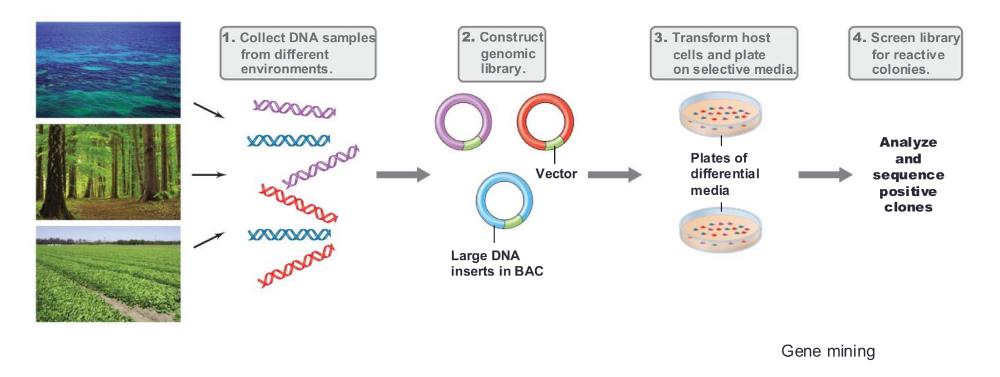
|
front 78 gene mining drawing | back 78 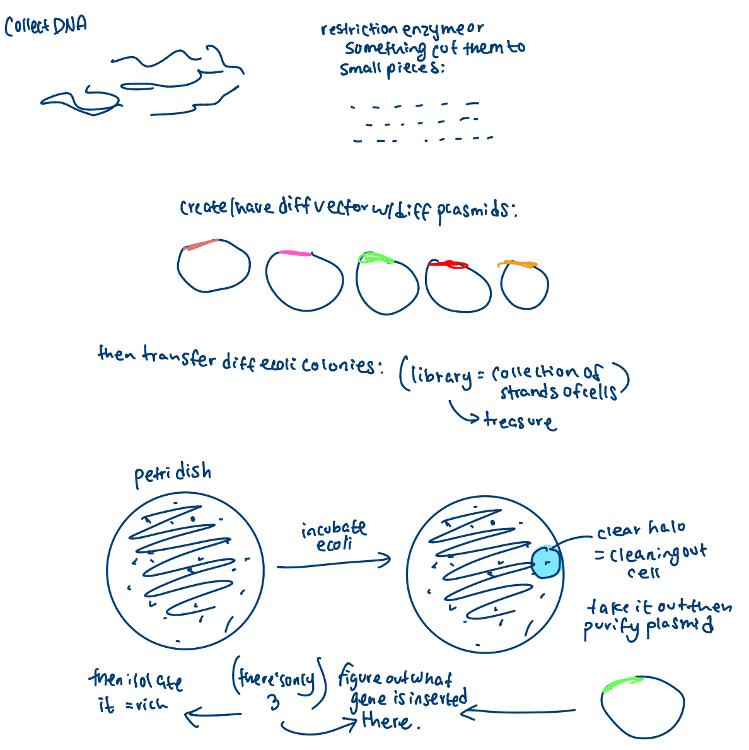 |
front 79 Engineering Metabolic Pathways | back 79 The production of small metabolites by genetic engineering typically involves multiple genes that must be expressed in a coordinated manner |
front 80 Pathway engineering | back 80 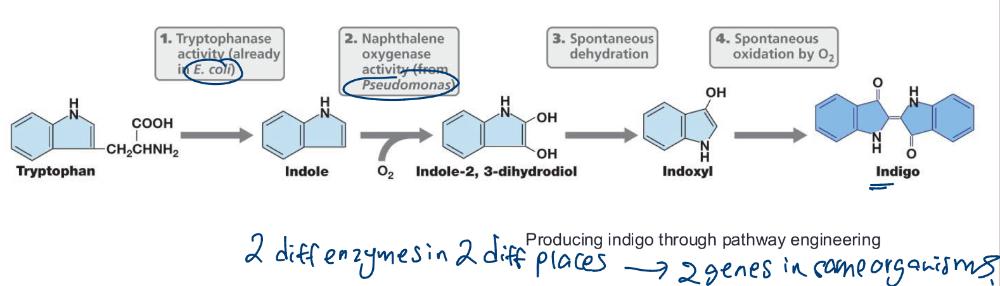 The process of assembling a new or improved biochemical pathway using genes from one or more organisms (e.g., indigo) |
front 81 Synthetic biology | back 81 using genetic engineering to create novel Examples
|
front 82 Biocontainment of Genetically Modified Organisms (GMO genome) | back 82
|
front 83 Recoding and control of genetically modified Escherichia coli | back 83 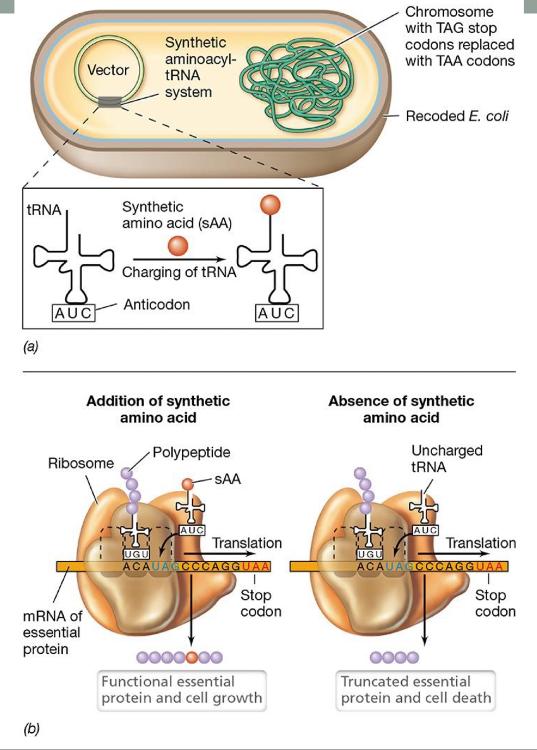 |