What is the proper order of the following events in the expression of a eukaryotic gene?
1. translation
2. RNA processing
3. transcription
4. modification of protein
- 4, 2, 3, 1
- 1, 2, 4, 3
- 1, 2, 3, 4
- 2, 3, 4, 1
- 3, 2, 1, 4
3, 2, 1, 4
Ex.
The proper order of the following events in the expression of a eukaryotic gene is: 3, 2, 1, 4.
3. transcription
2. RNA processing
1. translation
4. modification of protein
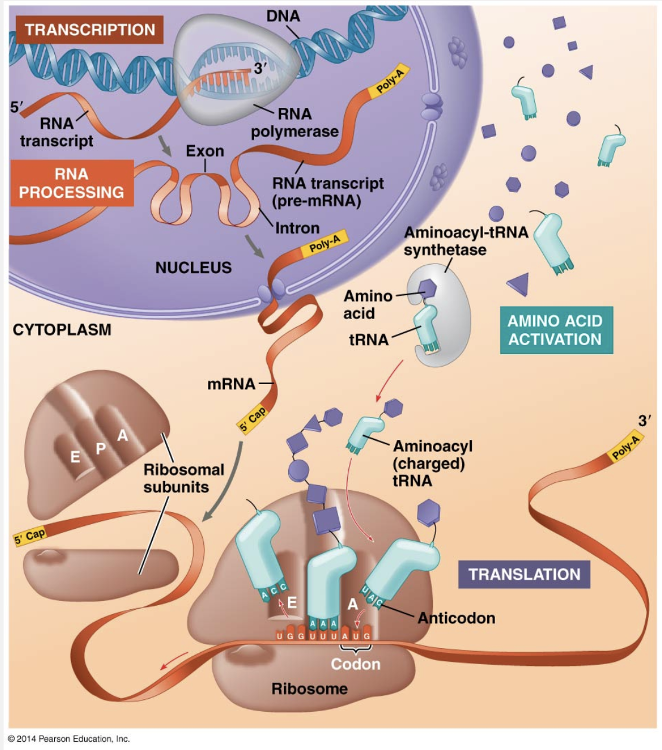
Transcription occurs in the nucleus, and mRNA is then transported to the cytoplasm, where translation occurs. But before eukaryotic RNA transcripts from protein-coding genes can leave the nucleus, they are modified in various ways to produce the final, functional mRNA. The transcription of a protein-coding eukaryotic gene results in pre-mRNA, and further processing yields the finished mRNA. The initial RNA transcript from any gene, including those specifying RNA that is not translated into protein, is more generally called a primary transcript. To summarize: Genes program protein synthesis via genetic messages in the form of messenger RNA. Put another way, cells are governed by a molecular chain of command with a directional flow of genetic information, shown here by arrows: DNA → RNA → PROTEIN.
“1, 2, 3, 4,” “4, 2, 3, 1,” “2, 3, 4, 1,” “1, 2, 4, 3” are all incorrect because transcription is the first step in gene expression.
Eukaryotic processing of the primary transcript includes __________.
- the addition of a 5ꞌ poly-A tail, a 3ꞌ cap, and the splicing out of exons
- the addition of a 5ꞌ cap, a 3ꞌ poly-A tail, the splicing out of introns, and a polyadenylation signal
- the addition of a 5ꞌ cap, a 3ꞌ poly-A tail, and the splicing out of exons
- the addition of a 5ꞌ cap, a 3ꞌ poly-A tail, and the splicing out of introns
- the addition of a 5ꞌ poly-A tail, a 3ꞌ cap, and the splicing out of introns
the addition of a 5ꞌ cap, a 3ꞌ poly-A tail, and the splicing out of introns
Ex.
Eukaryotic processing of the primary transcript includes the addition of a 5ꞌ cap, a 3ꞌ poly-A tail, and the splicing out of introns.
Each end of a pre-mRNA molecule is modified in a particular way. The 5ꞌ end, which is synthesized first, receives a 5ꞌ cap, a modified form of a guanine (G) nucleotide added onto the 5ꞌ end after transcription of the first 20–40 nucleotides. The 3ꞌ end of the pre-mRNA molecule is also modified before the mRNA exits the nucleus. Recall that the pre-mRNA is released soon after the polyadenylation signal, AAUAAA, is transcribed. At the 3ꞌ end, an enzyme then adds 50–250 more adenine (A) nucleotides, forming a poly-A tail. RNA processing in the eukaryotic nucleus involves the removal of large portions of the RNA molecule that is initially synthesized. This cut-and-paste job, called RNA splicing, is similar to editing a video. The average length of a transcription unit along a human DNA molecule is about 27,000 nucleotide pairs, so the primary RNA transcript is also that long. However, the average-sized protein of 400 amino acids requires only 1,200 nucleotides in RNA to code for it.
“The addition of a 5ꞌ cap, a 3ꞌ poly-A tail, and the splicing out of exons” is incorrect because exons are expressed segments of the mRNA and are not spliced out. “The addition of a 5ꞌ poly-A tail, a 3ꞌ cap, and the splicing out of introns” is incorrect because the 5ꞌ end of the mRNA has the cap and the 3ꞌ end has the poly-A tail. “The addition of a 5ꞌ poly-A tail, a 3ꞌ cap, and the splicing out of exons” is incorrect because the 5ꞌ end of the mRNA has the cap and the 3ꞌ end has the poly-A tail and because exons are expressed segments of the mRNA and are not spliced out. “The addition of a 5ꞌ cap, a 3ꞌ poly-A tail, the splicing out of introns, and a polyadenylation signal” is incorrect because the polyadenylation signal is coded for in the 3ꞌ untranslated region (UTR) of the gene.
By bombarding the fungus Neurospora crassa with X-rays, Beadle and Tatum were able to study __________ and characterize enzymes in a __________.
- phenotypic mutants; biochemical pathway
- wild-type mutants; biochemical pathway
- nutritional mutants; biochemical pathway
- phenotypic mutants; glycolysis
- nutritional mutants; glycolysis
nutritional mutants; biochemical pathway
Ex.
By bombarding the fungus Neurospora crassa with X-rays, Beadle and Tatum were able to study nutritional mutants and characterize enzymes in a biochemical pathway.
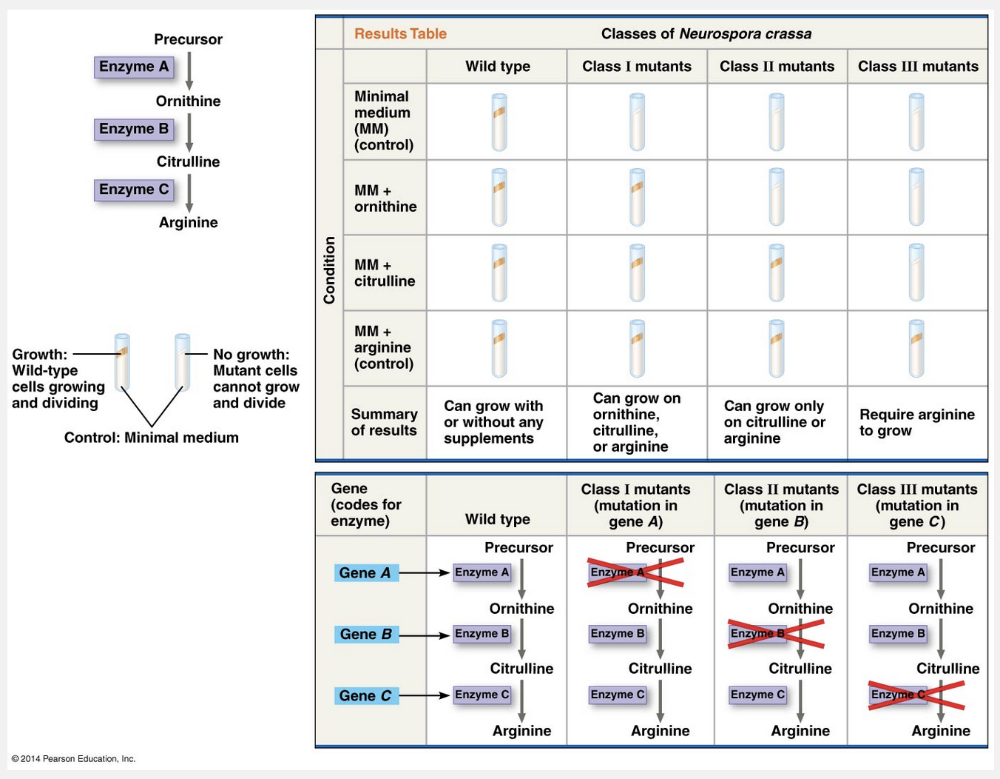
A breakthrough in demonstrating the relationship between genes and enzymes came a few years later at Stanford University, where George Beadle and Edward Tatum began working with a bread mold, Neurospora crassa. They bombarded Neurospora with X-rays, which had been shown in the 1920s to cause genetic changes, and then looked among the survivors for mutants that differed in their nutritional needs from those of the wild-type bread mold. They found that wild-type Neurospora has modest food requirements: It can grow in the laboratory on a simple solution of inorganic salts, glucose, and the vitamin biotin incorporated into agar, a support medium. From this minimal medium, the mold cells use their metabolic pathways to produce all the other molecules they need. Beadle and Tatum also identified mutants that could not survive on the minimal medium, apparently because they were unable to synthesize certain essential molecules from the minimal ingredients. To ensure the survival of these nutritional mutants, Beadle and Tatum allowed them to grow on a complete growth medium that consisted of the minimal medium supplemented with all 20 amino acids and a few other nutrients. The complete growth medium was able to support any mutant that couldn’t synthesize one of the supplements.
“Phenotypic mutants; biochemical pathway” is incorrect because the mutants that differed in their nutritional needs were from the wild-type bread mold. “Nutritional mutants; glycolysis” is incorrect because Beadle and Tatum studied amino acid biochemical pathways. “Phenotypic mutants; glycolysis” is incorrect because Beadle and Tatum studied amino acid biochemical pathways, and the mutants differed in their nutritional needs from those of the wild-type bread mold. “Wild-type mutants; biochemical pathway” is incorrect because the wild-type mold was not mutant, and the cells could use their metabolic pathways to produce all the other molecules they needed.
Bacteria can transcribe and translate human genes to produce functional human proteins because __________.
- eukaryotes do not really need a nucleus
- RNA has catalytic properties
- bacterial ribosomes and eukaryotic ribosomes are identical
- the genetic code is nearly universal
- bacterial and eukaryotic RNA polymerases are identical
the genetic code is nearly universal
Ex.
Bacteria can transcribe and translate human genes to produce functional human proteins because the genetic code is nearly universal.
The genetic code is nearly universal, shared by organisms from the simplest bacteria to the most complex plants and animals. The RNA codon CCG, for instance, is translated as the amino acid proline in all organisms whose genetic code has been examined. Exceptions to the universality of the genetic code include translation systems in which a few codons differ from the standard ones. Slight variations in the genetic code exist in certain unicellular eukaryotes and in the organelle genes of some species. Despite these exceptions, the evolutionary significance of the code’s near universality is clear. A language shared by all living things must have been operating very early in the history of life—early enough to be present in the common ancestor of all present-day organisms. A shared genetic vocabulary is a reminder of the kinship that bonds all life on Earth.
"Bacterial ribosomes and eukaryotic ribosomes are identical" is incorrect because they are not identical. Bacterial ribosomes are composed of 30S small subunit and 50S large subunit making a 70S ribosome whereas eukaryotic ribosomes are composed of 40S small subunit and 60S large subunit making an 80S ribosome. They also have different proteins and rRNAs.
"Eukaryotes do not really need a nucleus" is incorrect because all eukaryotic organisms have their genetic material in a nucleus.
"RNA has catalytic properties" is incorrect because although RNA has been shown to have catalytic properties, that is not why bacteria can transcribe and translate human genes.
"Bacterial and eukaryotic RNA polymerases are identical" is incorrect because bacteria and eukaryotic RNA polymerases are not the same, but they are capable of transcribing each other's DNA.
Genetic information of eukaryotic cells is transferred from the nucleus to the cytoplasm in the form of __________.
- lipids
- RNA
- proteins
- carbohydrates
RNA
Ex.
Genetic information of eukaryotic cells is transferred from the nucleus to the cytoplasm in the form of RNA.
In a eukaryotic cell, by contrast, the nuclear envelope separates transcription from translation in space and time. Transcription occurs in the nucleus, and mRNA is then transported to the cytoplasm, where translation occurs.
"Proteins” is not correct because in eukaryotes proteins are synthesized in the cytoplasm by ribosomes that read the messenger RNA. "DNA” is not correct because this molecule is never transferred to the cytoplasm for translation. "Carbohydrates” and "Lipids” are not correct because these molecules do not carry the genetic information.
The function of tRNA during protein synthesis is to __________.
- deliver amino acids to their proper site during protein synthesis
- guide ribosome subunits out of the nucleus through nuclear pores
- process mRNA
- transcribe mRNA
- attach mRNA to the small subunit of the ribosome
deliver amino acids to their proper site during protein synthesis
Ex.
The function of tRNA during protein synthesis is to deliver amino acids to their proper site during protein synthesis.
The message is a series of codons along an mRNA molecule, and the translator is called transfer RNA (tRNA). The function of tRNA is to transfer amino acids from the cytoplasmic pool of amino acids to a growing polypeptide in a ribosome. A cell keeps its cytoplasm stocked with all 20 amino acids, either by synthesizing them from other compounds or by taking them up from the surrounding solution.
“Guide ribosome subunits out of the nucleus through nuclear pores,” is not correct because the subunits are exported to the cytoplasm by the nuclear pores. “Attach mRNA to the small subunit of the ribosome” is incorrect because this occurs during the initiation stage prior to adding a tRNA. “Process mRNA” is incorrect because this is done by snRNPs. “Transcribe mRNA” is incorrect because this is done by RNA polymerase.
__________ is the synthesis of a polypeptide using information in the mRNA.
- The polypeptide hypothesis
- Transcription
- Translation
- Alternative splicing
- Splicing
Translation
Ex.
Translation is the synthesis of a polypeptide using information in the mRNA.
During this stage, there is a change in language: The cell must translate the nucleotide sequence of an mRNA molecule into the amino acid sequence of a polypeptide. The sites of translation are ribosomes, molecular complexes that facilitate the orderly linking of amino acids into polypeptide chains.
“Transcription” is incorrect because this is the synthesis of an mRNA molecule using the information in the DNA. “Splicing” is incorrect because this process occurs only in eukaryotic pre-mRNAs and only after the primary transcript has been synthesized. “Alternative splicing” is incorrect because this is a process that allows different mRNAs to be produced from the same primary transcript. “The polypeptide hypothesis” is incorrect because this is part of the restated Beadle and Tatum hypothesis of one gene–one enzyme.
Which of the following catalyzes the linkage between ribonucleotides to form RNA during gene expression?
- RNA polymerase
- RNA ligase
- Reverse transcriptase
- A ribozyme
- tRNA
RNA polymerase
Ex.
RNA polymerase catalyzes the linkage between ribonucleotides to form RNA during gene expression.
Transcription is the synthesis of RNA using information in the DNA. The two nucleic acids are written in different forms of the same language, and the information is simply transcribed, or “rewritten,” from DNA to RNA. Just as a DNA strand provides a template for making a new complementary strand during DNA replication, it also can serve as a template for assembling a complementary sequence of RNA nucleotides. For a protein-coding gene, the resulting RNA molecule is a faithful transcript of the gene’s protein-building instructions. An enzyme called an RNA polymerase pries the two strands of DNA apart and joins together RNA nucleotides complementary to the DNA template strand, thus elongating the RNA polynucleotide.
"RNA ligase" is incorrect because the enzyme that links ribonucleotides in gene expression is an RNA polymerase. "A ribozyme" is incorrect because this enzyme is involved in catalytic reaction reactions and not the addition of ribonucleotides. "Reverse transcriptase" is incorrect because this enzyme is a RNA dependent DNA polymerase and makes a DNA copy of RNA. "tRNA" is incorrect because this molecule carries the amino acid to the ribosome during translation. tRNA is an example of a RNA polymerase product.
Which of the following is a post-translational modification of a polypeptide?
- Removal of introns and splicing of exons
- Complementary base pairing of mRNA and tRNA in the ribosome
- The growing polypeptide signals the ribosome to attach to the ER
- Cleavage of a polypeptide into two or more chains
- Formation of a polysome that allows simultaneous formation of many polypeptides from one mRNA transcript
Cleavage of a polypeptide into two or more chains
Ex.
Cleavage of a polypeptide into two or more chains is an example of a post-translational modification of a polypeptide.
Additional steps—post-translational modifications—may be required before the protein can begin doing its particular job in the cell. Certain amino acids may be chemically modified by the attachment of sugars, lipids, phosphate groups, or other additions. Enzymes may remove one or more amino acids from the leading (amino) end of the polypeptide chain. In some cases, a polypeptide chain may be enzymatically cleaved into two or more pieces. For example, the protein insulin is first synthesized as a single polypeptide chain but becomes active only after an enzyme cuts out a central part of the chain, leaving a protein made up of two polypeptide chains connected by disulfide bridges. In other cases, two or more polypeptides that are synthesized separately may come together, becoming the subunits of a protein that has quaternary structure. A familiar example is hemoglobin.
“Removal of introns and splicing of exons” is incorrect because this is an example of a post-transcriptional modification. “Formation of a polysome that allows simultaneous formation of many polypeptides from one mRNA transcript” is incorrect because this is translation and occurs before the peptide is modified. “The growing polypeptide signals the ribosome to attach to the ER” is incorrect because this occurs near the leading end of the growing peptide chain. “Complementary base pairing of mRNA and tRNA in the ribosome” is incorrect because this occurs during translation and is not a post-translational modification.
What is a key difference in gene expression between eukaryotic and prokaryotic cells?
- In prokaryotes, proteins are assembled directly from DNA.
- In eukaryotic cells, transcribed RNA sequences function as termination signals.
- Prokaryotes do not contain ribosomes.
- In prokaryotic cells, the mRNA transcript is immediately available as mRNA without processing.
- RNA polymerases are involved only in initiation in eukaryotes.
In prokaryotic cells, the mRNA transcript is immediately available as mRNA without processing.
Ex.
The key difference in gene expression between eukaryotic and prokaryotic cells is that in prokaryotic cells, the mRNA transcript is immediately available as mRNA without processing.
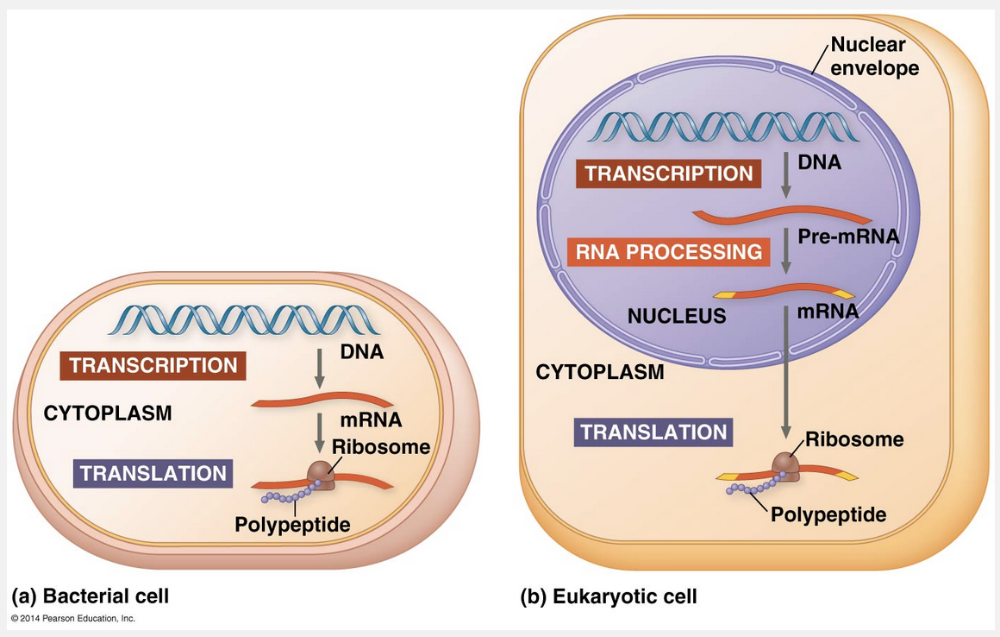
The most important differences between bacteria and eukaryotes with regard to gene expression arise from the bacterial cell’s lack of compartmental organization. Like a one-room workshop, a bacterial cell ensures a streamlined operation. In the absence of a nucleus, it can simultaneously transcribe and translate the same gene, and the newly made protein can quickly diffuse to its site of function. Most researchers suspect that transcription and translation are coupled like this in archaeal cells as well, because archaea lack a nuclear envelope. In contrast, the eukaryotic cell’s nuclear envelope segregates transcription from translation and provides a compartment for extensive RNA processing. This processing stage includes additional steps whose regulation can help coordinate the eukaryotic cell’s elaborate activities.
“In prokaryotes, proteins are assembled directly from DNA” is incorrect because proteins are always assembled from a mRNA transcript. “RNA polymerases are involved only in initiation in eukaryotes” is incorrect because both prokaryotes have an RNA polymerase. “In eukaryotic cells, transcribed RNA sequences function as termination signals” is incorrect because transcribed RNA sequences function as termination signals are found in prokaryotes too. “Prokaryotes do not contain ribosomes” is incorrect because all cells, prokaryotes, archaea, and eukaryotes have ribosomes.
The “triplet code” refers to the fact that _________.
- None of the listed responses is correct.
- three nucleotides code for a single amino acid
- three amino acids code for a single nucleotide
- the three-letter code for each amino acid is a triplet
- three sets of nucleotides are required for each amino acid

three nucleotides code for a single amino acid
Ex.
The “triplet code” refers to the fact that three nucleotides code for a single amino acid.
Triplets of nucleotide bases are the smallest units of uniform length that can code for all the amino acids. If each arrangement of three consecutive nucleotide bases specifies an amino acid, there can be 64 (that is, 43) possible code words—more than enough to specify all the amino acids. Experiments have verified that the flow of information from gene to protein is based on a triplet code: The genetic instructions for a polypeptide chain are written in the DNA as a series of nonoverlapping three-nucleotide words. The series of words in a gene is transcribed into a complementary series of nonoverlapping three-nucleotide words in mRNA, which is then translated into a chain of amino acids.
“Three amino acids code for a single nucleotide” is incorrect because three nucleotides form a codon that specifies a specific amino acid. “Three sets of nucleotides are required for each amino acid” is incorrect because only one set of three nucleotides is necessary to form a codon that specifies a specific amino acid. “The three-letter code for each amino acid is a triplet” is incorrect because this abbreviation does not represent a codon.
When RNA is being made, the RNA base _________ always pairs with the base __________ in DNA.
- U; A
- U; T
- T; G
- T; A
- A; U
U; A
Ex.
When RNA is being made, the RNA base U always pairs with the base A in DNA.
An mRNA molecule is complementary rather than identical to its DNA template because RNA nucleotides are assembled on the template according to base-pairing rules. The pairs are similar to those that form during DNA replication, except that U, the RNA substitute for T, pairs with A and the mRNA nucleotides contain ribose instead of deoxyribose. Like a new strand of DNA, the RNA molecule is synthesized in an antiparallel direction to the template strand of DNA.
"U; T” is incorrect because T always base-pairs with A. "T; G” is incorrect because G always base-pairs with C and T is never found in RNA molecules. "A; U” is incorrect because U is never found in DNA molecules. "T; A” is incorrect because T is never found in RNA molecules. This is an example of DNA-DNA base-pairing and not RNA-DNA base-pairing.
The TATA box is a sequence with a __________ that allows for the binding of __________ and __________.
- eukaryotic promoter; transcription factors; ribozymes
- prokaryotic promoter; translation factors; ribosome
- prokaryotic promoter; transcription factors; RNA polymerase II
- eukaryotic promoter; transcription factors; RNA polymerase II
- eukaryotic promoter; translation factors; ribosome
eukaryotic promoter; transcription factors; RNA polymerase II
Ex.
The TATA box is a eukaryotic promoter that allows for the binding of transcription factors and RNA polymerase II .
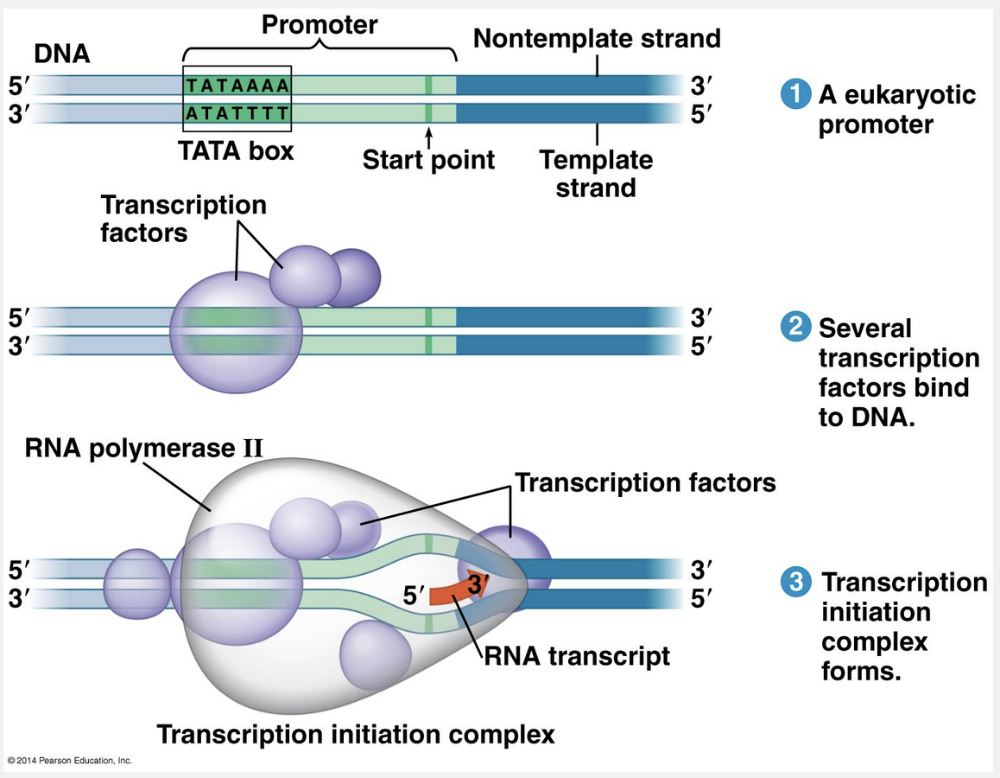
In eukaryotes, a collection of proteins called transcription factors mediate the binding of RNA polymerase and the initiation of transcription. Only after transcription factors are attached to the promoter does RNA polymerase II bind to it. The whole complex of transcription factors and RNA polymerase II bound to the promoter is called a transcription initiation complex.
This figure shows the roles of transcription factors and a crucial promoter DNA sequence called a TATA box in forming the initiation complex at a eukaryotic promoter.
“Prokaryotic promoter; transcription factors; RNA polymerase II,” “prokaryotic promoter; translation factors; ribosome,” “eukaryotic promoter; translation factors; ribosome,” and “eukaryotic promoter; transcription factors; ribozymes” are incorrect because the TATA box is a eukaryotic promoter involved in binding transcription factors and RNA polymerase II. Ribozymes are RNA molecules that function as enzymes.
Stop codons are unique because they __________.
- do not code for amino acids that allow a releasing factor to bind to the E site of the ribosome
- code for a specific amino acid that binds to a releasing factor
- do not code for amino acids that allow a releasing factor to bind to the P site of the ribosome
- do not code for amino acids but instead allow a releasing factor to bind to the A site of the ribosome
- code for releasing factors
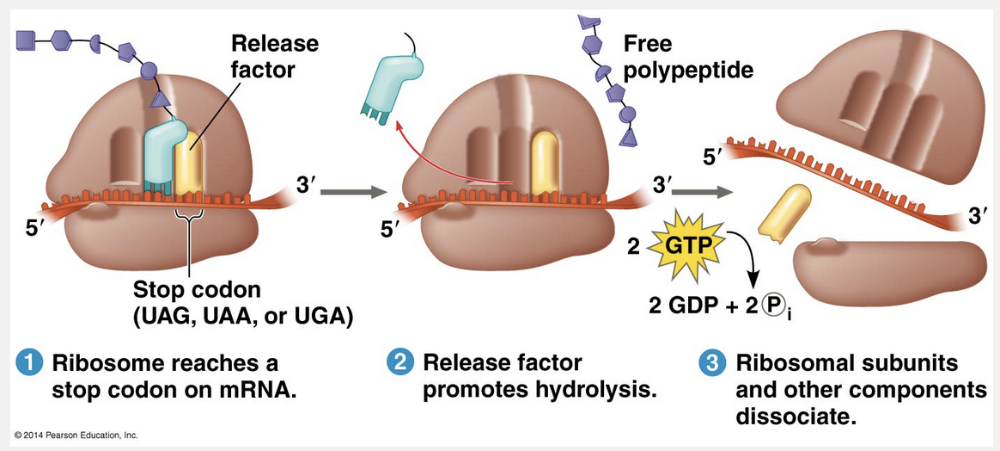
do not code for amino acids but instead allow a releasing factor to bind to the A site of the ribosome
Ex.
Stop codons are unique because they do not code for amino acids but instead allow a releasing factor to bind to the A site of the ribosome.
The final stage of translation is termination. Elongation continues until a stop codon in the mRNA reaches the A site of the ribosome. The nucleotide base triplets UAG, UAA, and UGA do not code for amino acids but instead act as signals to stop translation. A release factor, a protein shaped like an aminoacyl tRNA, binds directly to the stop codon in the A site. The release factor causes the addition of a water molecule, instead of an amino acid, to the polypeptide chain. (There are plenty of water molecules available in the aqueous cellular environment.) This reaction breaks (hydrolyzes) the bond between the completed polypeptide and the tRNA in the P site, releasing the polypeptide through the exit tunnel of the ribosome’s large subunit. The remainder of the translation assembly then comes apart in a multistep process, aided by other protein factors. Breakdown of the translation assembly requires the hydrolysis of two more GTP molecules.
“Do not code for amino acids that allow a releasing factor to bind to the P site of the ribosome” and “do not code for amino acids that allow a releasing factor to bind to the E site of the ribosome” are incorrect because releasing factors bind to the A site of the ribosome. “Code for a specific amino acid that binds to a releasing factor” is incorrect because stop codons do not code for amino acids that allow for a releasing factor to terminate translation. “Code for releasing factors” is incorrect because releasing factors are proteins that cause the termination of translation.
The type of point mutation that results in a premature stop codon is called a _________ mutation.
- missense
- silent
- inversion
- deletion
- nonsense
nonsense
Ex.
The type of mutation that results in a premature stop codon is called a nonsense mutation.
Substitution mutations are usually missense mutations; that is, the altered codon still codes for an amino acid and thus makes sense, although not necessarily the right sense. But a point mutation can also change a codon for an amino acid into a stop codon. This is called a nonsense mutation, and it causes translation to be terminated prematurely; the resulting polypeptide will be shorter than the polypeptide encoded by the normal gene. Nearly all nonsense mutations lead to nonfunctional proteins.
“Missense” is incorrect because this kind of mutation results in a substitution of one amino acid for another. “Silent” is incorrect because this kind of mutation has no observable effect on the phenotype. “Inversion” and “deletion” are incorrect because they are chromosomal mutations and not point mutations.
Polysomes may be defined as __________.
- groups of peroxisomes
- microfilaments and microtubules
- groups of ribosomes
- groups of lysosomes
- groups of chromosomes
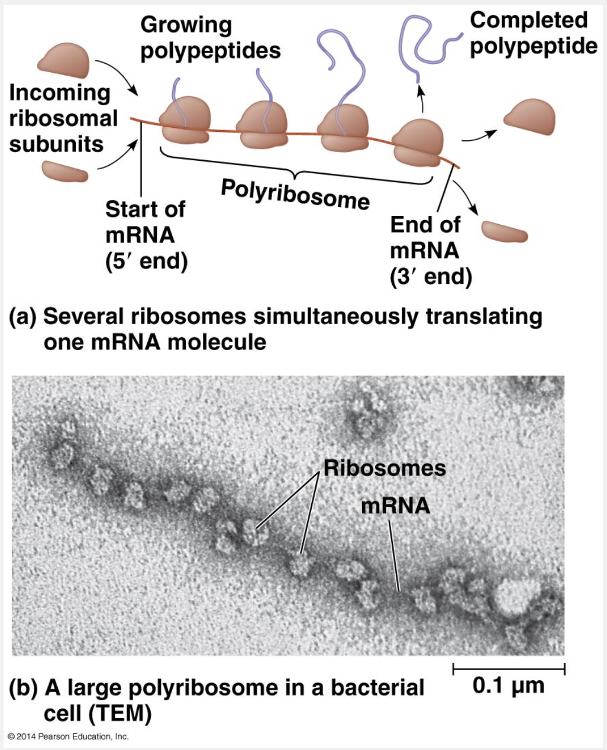
groups of ribosomes
Ex.
Polysomes may be defined as groups of ribosomes.
A single ribosome can make an average-sized polypeptide in less than a minute. Typically, however, multiple ribosomes translate an mRNA at the same time; that is, a single mRNA is used to make many copies of a polypeptide simultaneously. Once a ribosome is far enough past the start codon, a second ribosome can attach to the mRNA, eventually resulting in a number of ribosomes trailing along the mRNA. Such strings of ribosomes, called polyribosomes (or polysomes), can be seen with an electron microscope. Polyribosomes are found in both bacterial and eukaryotic cells. They enable a cell to make many copies of a polypeptide very quickly.
“Microfilaments and microtubules” is incorrect because these are cytoskeletal elements found in eukaryotic cells. “Groups of lysosomes,” “groups of peroxisomes,” and “groups of chromosomes” are incorrect because these are organelles found in eukaryotic cells.
Gene expression is __________.
- the process by which DNA directs the synthesis of proteins
- None of the listed responses is correct.
- the genetic makeup of an individual
- the way that some genes express themselves at different times of the day, giving an individual a new appearance
- the way that an individual appears
the process by which DNA directs the synthesis of proteins
Ex.
Gene expression is the process by which DNA directs the synthesis of proteins.
The DNA inherited by an organism leads to specific traits because DNA dictates the synthesis of proteins or, in some cases, just RNA molecules. In other words, proteins are the link between genotype and phenotype. The expression of genes that code for proteins includes two stages: transcription and translation.
“The genetic makeup of an individual” and “the way that an individual appears” are incorrect because gene expression is the process by which DNA directs the synthesis of proteins (or, in some cases, just RNAs) and is the link between genotype and phenotype. “The way that some genes express themselves at different times of the day, giving an individual a new appearance,” is incorrect because gene expression does not change an individual’s phenotype at different times of the day.
One strand of a DNA molecule has the following sequence: 3-AGTACAAACTATCCACCGTC-5.
In order for transcription to occur in that strand, there would have to be a specific recognition sequence, called a(n) __________, to the left of the DNA sequence indicated.
- promoter
- intron
- AUG codon
- exon
- centromere
promoter
Ex.
One strand of a DNA molecule has the following sequence: 3'-AGTACAAACTATCCACCGTC-5'. In order for transcription to occur in that strand, there would have to be a specific recognition sequence, called a promoter, to the left of the DNA sequence indicated.
Specific sequences of nucleotides along the DNA mark where transcription of a gene begins. The DNA sequence where RNA polymerase attaches and initiates transcription is known as the promoter. The promoter of a gene includes within it the transcription start point (the nucleotide where RNA synthesis actually begins) and typically extends several dozen or more nucleotide pairs upstream from the start point. RNA polymerase binds in a precise location and orientation on the promoter, therefore determining where transcription starts and which of the two strands of the DNA helix is used as the template.
"Centromere" is incorrect because this is a structure that is part of a eukaryotic chromosome. "Intron" is incorrect because this is a region of the pre-mRNA that is spliced out during RNA processing. "Exon" is incorrect because this is the region of the pre-mRNA that is retaining during RNA processing and contains the codons that code for the specific amino acids of the gene. "AUG codon" is incorrect because this codon codes for the amino acid methionine and is also the start codon during translation initiation.
The bonds that hold tRNA molecules in the correct three-dimensional shape are __________.
- covalent bonds
- ionic bonds
- hydrogen bonds
- peptide linkages
- hydrophobic interactions
hydrogen bonds
Ex.
The bonds that hold tRNA molecules in the correct three-dimensional shape are hydrogen bonds.
A tRNA molecule consists of a single RNA strand that is only about 80 nucleotides long (compared to hundreds of nucleotides for most mRNA molecules). Because of the presence of complementary stretches of nucleotide bases that can hydrogen bond to each other, this single strand can fold back upon itself and form a molecule with a three-dimensional structure. Flattened into one plane to clarify this base pairing, a tRNA molecule looks like a cloverleaf.
“Peptide linkages” is incorrect because these bonds amino acids. “Hydrophobic interactions” is incorrect because these interactions occur in protein folding when a specific amino acid is hydrophobic caused a region of the protein to fold in on itself away from the aqueous environment. “Covalent bonds” is incorrect because these bond form between atoms that share a hybrid orbital. “Ionic bonds” is incorrect because these bonds form when there are charge attractions such is with sodium and chlorine to form the salt NaCl.
RNA molecules that function as enzymes are called __________.
- aminoacyl-RNA synthetases
- polysomes
- transfer RNAs
- ribozymes
- RNA polymerases
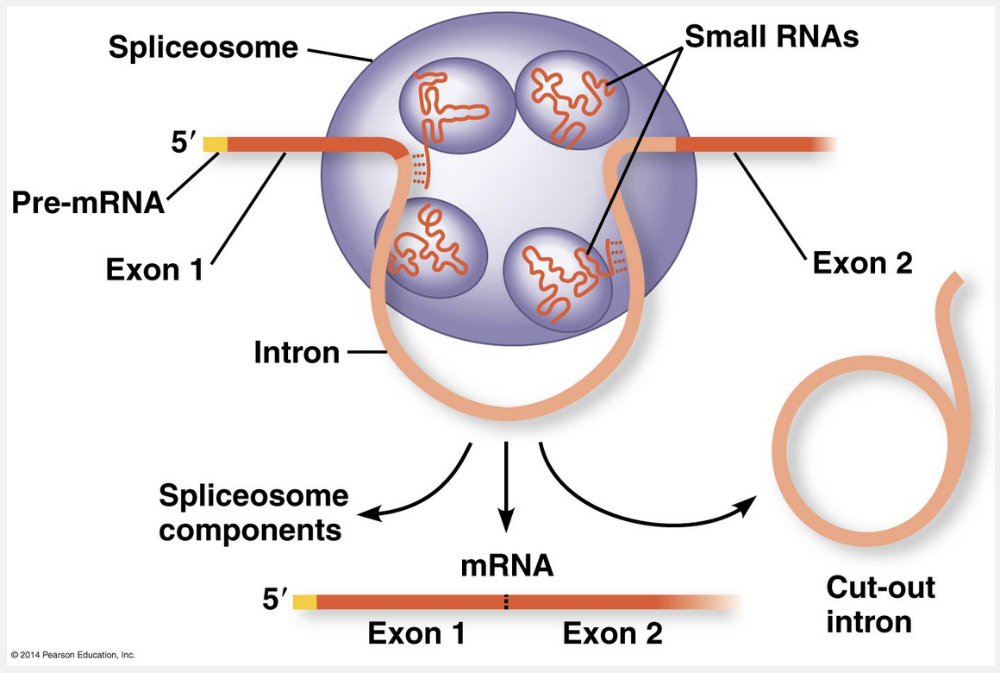
ribozymes
Ex.
RNA molecules that function as enzymes are called ribozymes.
The idea of a catalytic role for the RNAs in the spliceosome arose from the discovery of ribozymes, RNA molecules that function as enzymes. In some organisms, RNA splicing can occur without proteins or even additional RNA molecules: The intron RNA functions as a ribozyme and catalyzes its own excision! For example, in the ciliate protist Tetrahymena, self-splicing occurs in the production of ribosomal RNA (rRNA), a component of the organism’s ribosomes. The pre-rRNA actually removes its own introns. The discovery of ribozymes rendered obsolete the idea that all biological catalysts are proteins.
“RNA polymerases” is incorrect because these are proteins that function as an enzyme. “Transfer RNAs” is incorrect because these RNA molecules bring the correct amino acid to the ribosome. “Polysomes” is incorrect because they are strings of ribosomes. “Aminoacyl-RNA synthetases” is incorrect because these enzymes attach the correct amino acid to its specific tRNA.
An exception to the one gene–one enzyme hypothesis is __________.
- that all genes code for enzymes that produce structural proteins such as keratin
- None of the listed responses is correct.
- that not all amino acids code for enzymes; some amino acids code for structural proteins such as keratin
- that not all genes code for enzymes; some genes code for structural proteins such as keratin
- that the genetic code is for enzymes, while the structural code is for structural proteins such as keratin
that not all genes code for enzymes; some genes code for structural proteins such as keratin
Ex.
An exception to the one gene–one enzyme hypothesis is that not all genes code for enzymes; some genes code for structural proteins such as keratin.
As researchers learned more about proteins, they made revisions to the one gene–one enzyme hypothesis. First of all, not all proteins are enzymes. Keratin, the structural protein of animal hair, and the hormone insulin are two examples of nonenzyme proteins. Because proteins that are not enzymes are nevertheless gene products, molecular biologists began to think in terms of one gene–one protein. However, many proteins are constructed from two or more different polypeptide chains, and each polypeptide is specified by its own gene. For example, hemoglobin, the oxygen-transporting protein of vertebrate red blood cells, contains two kinds of polypeptides, and thus two genes code for this protein. Beadle and Tatum’s idea was therefore restated as the one gene–one polypeptide hypothesis.
“That not all amino acids code for enzymes; some amino acids code for structural proteins such as keratin” is incorrect because genes code for proteins. “That all genes code for enzymes that produce structural proteins such as keratin” is incorrect because some genes code for RNA molecules. “That the genetic code is for enzymes, while the structural code is for structural proteins such as keratin” is incorrect because there is only a universal genetic code and no structural code.
Which of the following best describes the arrangement of genetic information in a DNA molecule?
- The three-nucleotide words of a gene are arranged in a nonoverlapping series on the DNA template strand.
- By overlapping the three-nucleotide words of a gene, the amount of information a DNA molecule can hold is maximized.
- A gene is composed of overlapping, three-nucleotide words on a template strand of DNA.
- By analyzing the linear order of amino acids in a polypeptide, the exact order of the three-nucleotide words of a gene arranged on the template strand of DNA can be determined.
- The three-nucleotide words of a gene are serially arranged on both strands of DNA at a specific locus.
The three-nucleotide words of a gene are arranged in a nonoverlapping series on the DNA template strand.
Ex.
The response the three-nucleotide words of a gene are arranged in a nonoverlapping series on the DNA template strand best describes the arrangement of genetic information in a DNA molecule.
The genetic instructions for a polypeptide chain are written in the DNA as a series of nonoverlapping, three-nucleotide words. The series of words in a gene is transcribed into a complementary series of nonoverlapping, three-nucleotide words in mRNA, which is then translated into a chain of amino acids.
“A gene is composed of overlapping, three-nucleotide words on a template strand of DNA” is incorrect because although it is written as three-nucleotide words it is not overlapping.
“By analyzing the linear order of amino acids in a polypeptide, the exact order of the three-nucleotide words of a gene arranged on the template strand of DNA can be determined” is incorrect because the amino acid sequence is identical to the coding strand of the DNA except that T is replaced with U. During transcription, the gene determines the sequence of nucleotide bases along the length of the RNA molecule that is being synthesized. For each gene, only one of the two DNA strands is transcribed. This strand is called the template strand because it provides the pattern, or template, for the sequence of nucleotides in an RNA transcript.
“By overlapping the three-nucleotide words of a gene, the amount of information a DNA molecule can hold is maximized” is incorrect because the instructions are written as a DNA series of nonoverlapping, three nucleotide words.
“The three-nucleotide words of a gene are serially arranged on both strands of DNA at a specific locus” is incorrect because the 5’ strand is the coding strand and is identical to the transcript except that Us are used in RNA and the 3’ strand is called the template strand and provide the pattern for the sequence of nucleotides in the RNA transcript.
Insertions and deletions are called __________ mutations.
- missense
- frameshift
- silent
- nucleotide-pair substitution
- nonsense
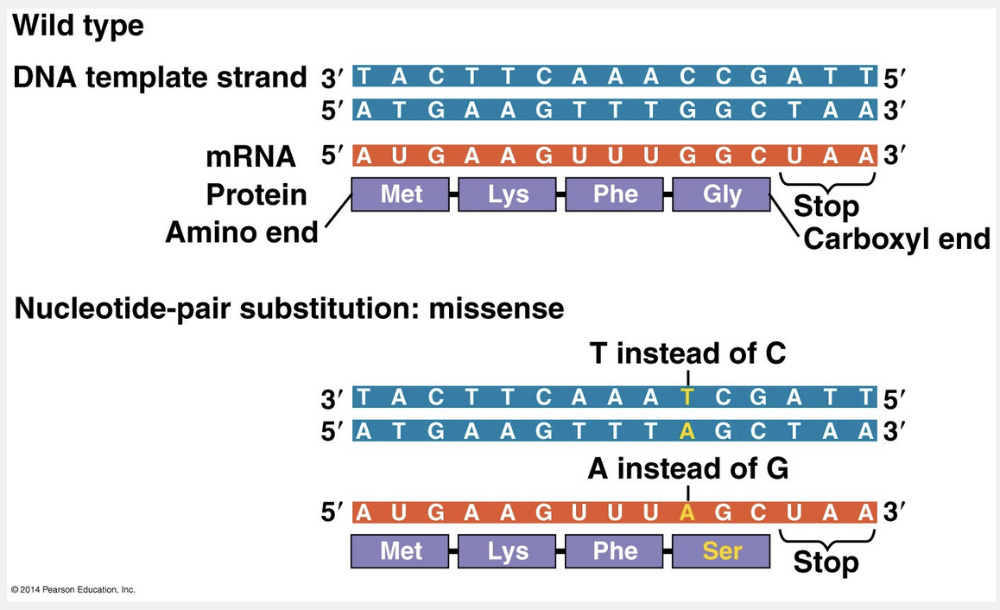
frameshift
Ex.
Insertions and deletions are called frameshift mutations.
Insertions and deletions are additions or losses of nucleotide pairs in a gene. These mutations have a disastrous effect on the resulting protein more often than substitutions do. Insertion or deletion of nucleotides may alter the reading frame of the genetic message, the triplet grouping of nucleotides on the mRNA that is read during translation. Such a mutation occurs whenever the number of nucleotides inserted or deleted is not a multiple of three. All nucleotides downstream of the deletion or insertion will be improperly grouped into codons; the result will be extensive missense, usually ending sooner or later in nonsense and premature termination. Unless the frameshift is very near the end of the gene, the protein is almost certain to be nonfunctional.
“Missense” is incorrect because this kind of mutation results in a substitution of one amino acid for another. “Nonsense” is incorrect because this kind of mutation changes a codon for an amino acid to a stop codon. “Silent” is incorrect because this kind of mutation has no observable effect on the phenotype. “Nucleotide-pair substitution” is incorrect because it is the replacement of one nucleotide and its partner with another pair of nucleotides.
Because the bacterial cell’s DNA is not surrounded by a nuclear envelope, __________ occur(s).
- coupled transcription and translation
- alternative splicing
- coupled splicing and tailing of the message
- segregated splicing and tailing of the message
- segregated transcription and translation
coupled transcription and translation
Ex.
Because the bacterial cell’s DNA is not surrounded by a nuclear envelope, coupled transcription and translation occur.
Another way both bacteria and eukaryotes augment the number of copies of a polypeptide is by transcribing multiple mRNAs from the same gene, as we mentioned earlier. However, the coordination of the two processes—transcription and translation—differs in the two groups. The most important differences between bacteria and eukaryotes arise from the bacterial cell’s lack of compartmental organization. Like a one-room workshop, a bacterial cell ensures a streamlined operation by coupling the two processes. In the absence of a nucleus, it can simultaneously transcribe and translate the same gene, and the newly made protein can quickly diffuse to its site of function.
“Coupled splicing and tailing of the message, “alternative splicing,” and “segregated transcription and translation” are incorrect because these processes occur only in a eukaryotic cell. “Segregated splicing and tailing of the message” is incorrect because these processes occur only in the eukaryotic cytosol.
Who formulated the one gene–one enzyme hypothesis?
- Beadle and Tatum
- Watson and Crick
- Hershey and Chase
- None of the listed responses is correct.
- Franklin
Beadle and Tatum
Ex.
Beadle and Tatum formulated the one gene-one enzyme hypothesis.
Beadle and Tatum worked with a bread mold called Neuropora that they had bombarded with X-rays resulting in mutants. Because each mutant was defective in a single gene, Beadle and Tatum saw that, taken together, the collected results provided strong support for a working hypothesis they had proposed earlier. The one gene–one enzyme hypothesis, as they dubbed it, states that the function of a gene is to dictate the production of a specific enzyme. Further support for this hypothesis came from experiments that identified the specific enzymes lacking in the mutants. Beadle and Tatum shared a Nobel Prize in 1958 for “their discovery that genes act by regulating definite chemical events” (in the words of the Nobel committee).
Watson and Crick is incorrect because they studied the structure of the DNA molecule by making models.
Hershey and Chase is incorrect because they performed experiments showing that DNA is the genetic material of a bacteriophage known as T2.
Franklin is incorrect because she was an X-ray crystallographer who studied the structure of DNA and produced the famous diffraction image of double stranded DNA.
How many nucleotides are needed to code for a protein with 450 amino acids?
- At least 300
- At least 1,350
- At least 900
- At least 450
- At least 150
At least 1,350
Ex.
The number of nucleotides that are needed to code for a protein with 450 amino acids is at least 1,350.
During translation, the sequence of codons along an mRNA molecule is decoded, or translated, into a sequence of amino acids making up a polypeptide chain. The codons are read by the translation machinery in the 5' → 3' direction along the mRNA. Each codon specifies which one of the 20 amino acids will be incorporated at the corresponding position along a polypeptide. Because codons are nucleotide triplets, the number of nucleotides making up a genetic message must be three times the number of amino acids in the protein product. For example, it takes 300 nucleotides along an mRNA strand to code for the amino acids in a polypeptide that is 100 amino acids long.
"At least 150" is incorrect because the number of bases needed to make a 450 amino acid protein is not one third of the number of amino acids but three times the number of amino acids in a protein. "At least 300" is not correct because this number is on two-thirds the number of amino acids in this protein. "At least 450" is incorrect because this number is equal to the number of amino acids. "At least 900" is incorrect because this number is only twice the number of amino acids in the protein and the correct answer is three times as much or 450 x 3 = 1,350.
The structures called snRNPs are __________.
- involved in the removal of exons from DNA
- part of a spliceosome
- a critical component of the initiation complex
- All of the listed responses are correct.
- a type of specialized carbohydrate
part of a spliceosome
Ex.
The structures called snRNPs are part of a spliceosome.
Particles called small nuclear ribonucleoproteins, abbreviated snRNPs (pronounced “snurps”), recognize these splice sites. As the full name implies, snRNPs are located in the cell nucleus and are composed of RNA and protein molecules. The RNA in a snRNP particle is called a small nuclear RNA (snRNA); each snRNA molecule is about 150 nucleotides long. Several different snRNPs join with additional proteins to form an even larger assembly called a spliceosome, which is almost as big as a ribosome. The spliceosome interacts with certain sites along an intron, releasing the intron, which is rapidly degraded, and joining together the two exons that flanked the intron.
"Involved in the removal of exons from DNA" is incorrect because snRNPs remove introns and splice exons together. "A type of specialized carbohydrate" is incorrect because snRNPs are composed of RNA and protein molecules. "A critical component of the initiation complex" is incorrect because snRNPs are involved in RNA processing.
The sickle-cell β-globin mutation is an example of a __________.
- missense mutation
- nonsense mutation
- silent mutation
- base deletion
- pointless mutation
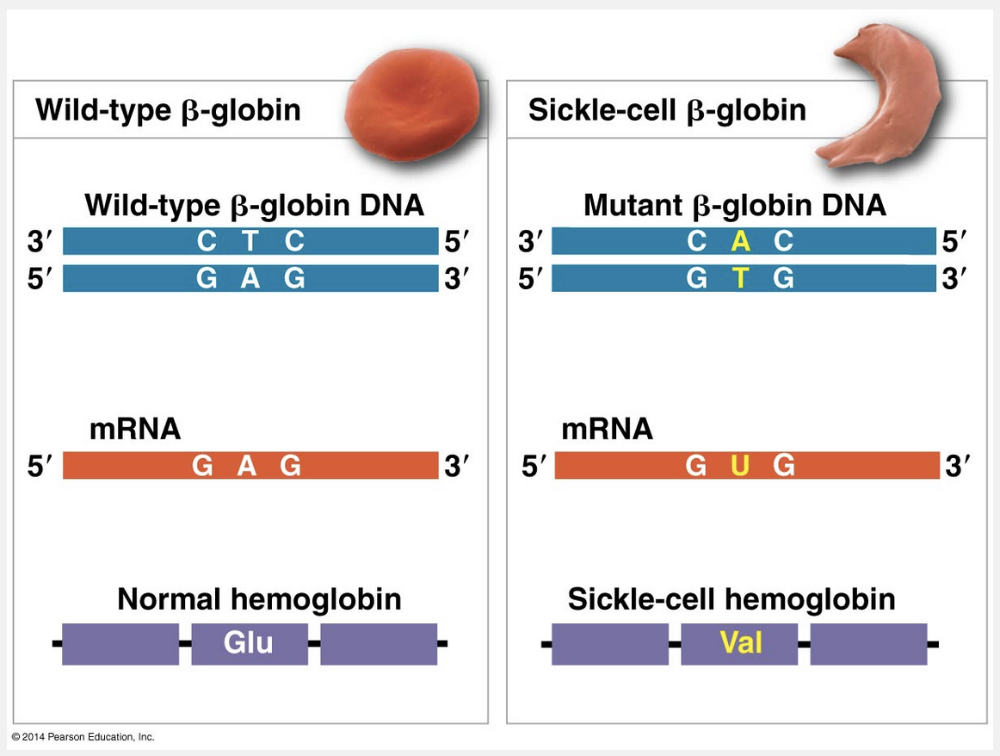
missense mutation
Ex.
The sickle-cell β-globin mutation is an example of a missense mutation.
The change of a single nucleotide in the DNA’s template strand leads to the production of an abnormal protein. In individuals who are homozygous for the mutant allele, the sickling of red blood cells caused by the altered hemoglobin produces the multiple symptoms associated with sickle-cell disease. Substitutions that change one amino acid to another are called missense mutations. Such a mutation may have little effect on the protein: The new amino acid may have properties similar to those of the amino acid it replaces, or it may be in a region of the protein where the exact sequence of amino acids is not essential to the protein’s function. Thus, the nucleotide-pair substitutions of greatest interest are those that cause a major change in a protein. The alteration of a single amino acid in a crucial area of a protein—such as in the part of the β-globin subunit of hemoglobin shown below—can significantly alter protein activity.
“Pointless mutation” is incorrect because there is no such term in genetics. “Silent mutation” is incorrect because the point mutation in sickle-cell disease results in a missense mutation and the substitution of the amino acid Glu (glutamic acid) for the amino acid Val (valine). “Nonsense mutation” is incorrect because mutation changes a codon for an amino acid to a stop codon. “Base deletion” is incorrect because such a deletion results in a frameshift mutation.
In eukaryotic cells, a __________ by a __________ targets a growing peptide to the endoplasmic reticulum.
- ribosome; signal-recognition particle
- signal-recognition particle; ribosome
- signal peptide; signal-recognition particle
- signal peptide; polyribosome
- signal-recognition particle; signal peptide
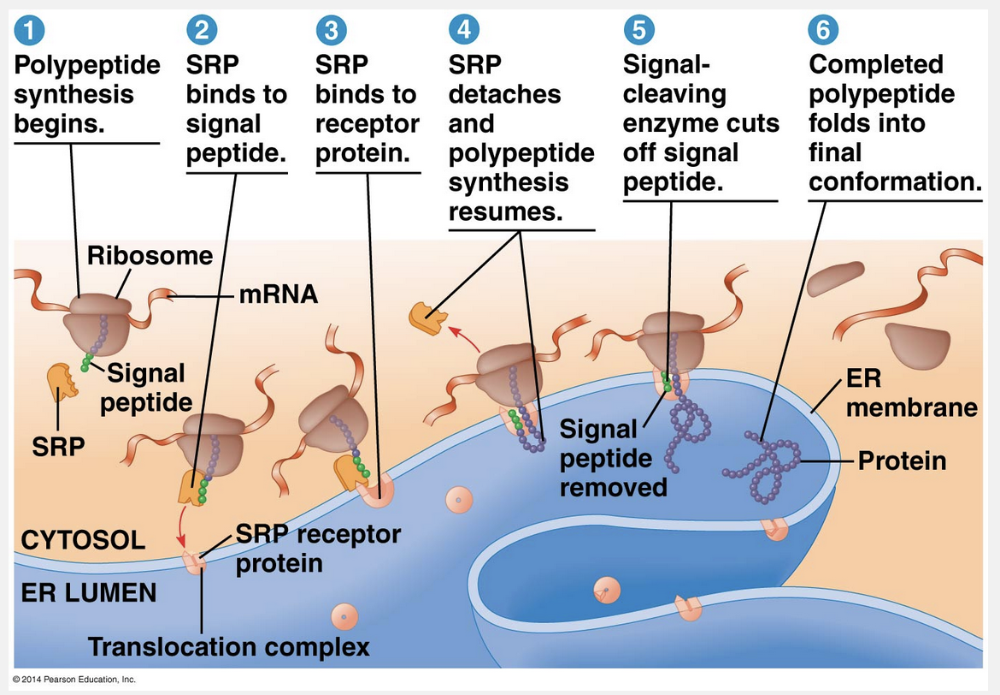
signal peptide; signal-recognition particle
Ex.
In eukaryotic cells, a signal peptide by a signal-recognition particle targets a growing peptide to the endoplasmic reticulum.
What determines whether a ribosome is free in the cytosol or bound to rough ER? Polypeptide synthesis always begins in the cytosol as a free ribosome starts to translate an mRNA molecule. There the process continues to completion, unless the growing polypeptide itself cues the ribosome to attach to the ER. The polypeptides of proteins destined for the endomembrane system or for secretion are marked by a signal peptide, which targets the protein to the ER. The signal peptide, a sequence of about 20 amino acids at or near the leading end (N-terminus) of the polypeptide, is recognized as it emerges from the ribosome by a protein-RNA complex called a signal-recognition particle (SRP). This particle functions as an escort that brings the ribosome to a receptor protein built into the ER membrane. The receptor is part of a multiprotein translocation complex. Polypeptide synthesis continues there, and the growing polypeptide snakes across the membrane into the ER lumen via a protein pore. The signal peptide is usually removed by an enzyme. The rest of the completed polypeptide, if it is to be secreted from the cell, is released into solution within the ER lumen.
“Ribosome; signal-recognition particle,” “signal-recognition particle; signal peptide,” “signal-recognition particle; ribosome,” and “signal peptide; polyribosome” are incorrect because the signal peptide of the growing polypeptide chain is targeted by a signal-recognition particle that directs the ribosome, mRNA, and peptide chain to the endoplasmic reticulum.
__________ is the synthesis of RNA using information in the DNA.
- Splicing
- Transcription
- Alternative splicing
- Translation
- The polypeptide hypothesis

Transcription
Ex.
Transcription is the synthesis of RNA using information in the DNA.
The two nucleic acids are written in different forms of the same language, and the information is simply transcribed, or “rewritten,” from DNA to RNA. Just as a DNA strand provides a template for making a new complementary strand during DNA replication, it also can serve as a template for assembling a complementary sequence of RNA nucleotides. For a protein-coding gene, the resulting RNA molecule is a faithful transcript of the gene’s protein-building instructions. This type of RNA molecule is called messenger RNA (mRNA) because it carries a genetic message from the DNA to the protein-synthesizing machinery of the cell. (Transcription is the general term for the synthesis of any kind of RNA on a DNA template.)
“Translation” is incorrect because this is the synthesis of a polypeptide using the information in the messenger RNA (mRNA). “Splicing” is incorrect because this process occurs only in eukaryotic pre-mRNAs and only after the primary transcript has been synthesized. “Alternative splicing” is incorrect because this is a process that allows different mRNAs to be produced from the same primary transcript. “The polypeptide hypothesis” is incorrect because this is part of the restated Beadle and Tatum hypothesis of one gene–one enzyme.