Genetic variation
refers to differences between members of
the same species or
those of different species
- allelic variation: due to differences in particular genes
- chromosomes: vary in structure (chromosome segments can be deleted, duplicated, and/or rearranged) & number (# individual chromosomes may vary (3 not 2) or numbers of chromosome sets (4 sets not the usual 2)
Cytogenetics
field of genetics that involves the microscopic examination of chromosomes
Cytogeneticist
examines the chromosomal composition of a particular cell or organism
- allows detection of individuals with abnormal chromosome number or structure
- provides a way to distinguish b/w species (human vs fruit fly vs corn)
Cytogeneticists use three main features to identify and classify chromosomes:
1. Location of centromere
2. Size
3. Banding patterns
N: these features are all seen in a karyotype
Karyotype
an individual's complete set of chromosomes
N: seen by micrograph
G banding
Chromosomes are exposed to the dye Giemsa
- shows dark and light bands
- 300 G bands seen in metaphase, 800 G bands in prometaphase
The G banding pattern is useful because:
- distinguishes individual chromosomes from each other
- detects changes in chromosome structure
reveals evolutionary relationships among the chromosomes of closely related species
Comparison of centromeric locations:
metacentric, submetacentric, acrocentric, telocentric
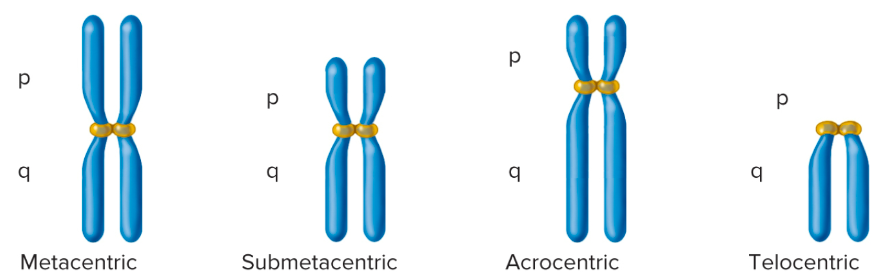
See image
2 Two primary ways chromosome structures can be altered
1. Total amount of genetic material in the chromosome can change
- deletion (deficiency): loss of a chromosomal segment
- duplication: repetition of a chromosomal segment compared to a normal chromosome
2. Total amount of genetic material remains the same but is rearranged
- inversion: change in the direction of part of the genetic material along a single chromosome
- translocation: a segment of one chromosome becomes attached to a different chromosome
Simple translocation
A piece of a chromosome is attached to another chromosome
Reciprocal translocations
Two different types of chromosomes exchange pieces, producing two abnormal chromosomes with translocations.
Two non-homologous chromosomes exchange genetic
material
- Reciprocal translocations arise from two different
mechanisms
- Chromosomal breakage and DNA repair
- Abnormal crossovers
Leads to a rearrangement of the genetic material, not a change in
the total amount
- Thus aka balanced translocations
- like inversions, are usually without phenotypic
consequences
- In a few cases, they can result in a breakpoint
effect or a position effect
Unlike reciprocal translocations, some translocations alter the
total amount of genetic material
- Called unbalanced
translocations, when genetic material is duplicated and/or
deleted
- Unbalanced translocations are associated with
phenotypic abnormalities or even lethality
Example:
Familial Down Syndrome
• In this condition, the majority of chromosome 21 is attached
to chromosome 14
- Offspring may have three copies of genes found
on a large segment of chromosome 21
Terminal deletion
a deletion that occurs towards the end of a chromosome; during chromosomal deletion, the chromosome breaks in two pieces and the fragment w/o the centromere is lost
Interstitial deletion
a deletion that occurs from the interior of a chromosome; chromosome breaks in two places, central fragment is lost, two outer pieces reattach
Phenotypic consequences of deletion depends on
- size of deletion
- chromosomal material deleted (are they vital)
Cri-du-chat syndrome
detrimental condition caused by chromosomal deletion in the short arm of chromosome 5
Duplication
chromosomal duplication is usually caused by abnormal events during recombination
- repetitive sequences (shown in red): can cause misalignment between homologous chromosomes
- If a crossover occurs, nonallelic homologous recombination results
Phenotypic consequences of duplications
- tend to be correlated to size (bigger piece, more likely to have effect)
- end to have less harmful effects the deletions of comparable size
Gene family
consists of two or more genes in a single species that are derived from the same ancestral gene.
N: The majority of small chromosomal duplications have
no
phenotypic effect but are vital because they provide the raw
material for the addition of genes to a species. This
can ultimately lead to the formation of gene families.
Paralogs
homologous genes within a single species
N: Homolog is the umbrella term for a genes that share origin. Orthologs are two genes in two different species that share a common ancestor, while paralogs are two genes in the same genome that are a product of a gene duplication event of the original gene.
Copy Number Variation (CNV)
segment of DNA that varies in copy number among members of the same species
- may be missing a particular gene
- may be a duplication
Possible mechanisms of CNV (Copy Number Variation)
- nonallelic homologous recombination
- proliferation of transposable elements (mobile DNA sequences capable of replicating themselves within genomes independently of the host cell DNA)
- errors in DNA replication
CNV associated w some human diseases
-Schizophrenia
- Autism and some certain learning disabilities
- Susceptibility to infectious diseases
- Cancer
Pericentric inversion
the inverted segment includes the centromere (with both arms involved)
Paracentric inversion
the two breaks appear on the same side of the centromere (in the same arm).
Break point effect:
An inversion break point occurs within a vital gene, thereby separating it into 2 nonfunctional parts
Position effect:
A gene is repositioned in a way that alters its gene expression
Inversion heterozygote
Individuals with one copy of a normal chromosome and one copy of an
inverted chromosome
- may have a high probability of producing
gametes that are abnormal in their total genetic contentN: The
abnormality is due to crossing over within the inverted segment
Inversion Heterozygotes:
During meiosis I, pairs of homologous sister chromatids synapse with
each other
- For the normal and inversion chromosome to synapse
properly, an inversion loop must form
- If a crossover occurs
within the inversion loop, highly abnormal chromosomes are produced
Robertsonian Translocation
Familial Down Syndrome is an example of
Robertsonian
translocation
This translocation occurs from
the following:
- Breaks occur near the centromeres of two
non-homologous acrocentric chromosomes
- The small acentric
fragments are lost
- The larger fragments fuse at their
centromeric regions to form a single chromosome which is metacentric
or submetacentric
Most common rearrangement in humans
-
Approximately one in 900 births
Euploidy
Variation in the number of complete sets of chromosome
Aneuploidy
Variation in the number of particular chromosomes within a set
- trisomic: organism has 3 copies of a chromosome instead of 2
- monosomic: 1 copy instead of 2
Commonly causes an abnormal phenotype
- It leads to an
imbalance in the amount of gene products
- Three copies of a gene
will lead to 150% production
- A single chromosome can have
hundreds or even
thousands of genes... Large excess of gene products
Imbalance of gene products produce individuals that are less likely to survive than a euploid individual.
Alterations in chromosome number occur frequently during gamete formation.
The autosomal aneuploidies compatible with survival are trisomies 13, 18 and 21. Aneuploidies involving sex chromosomes generally have less severe effects than those of autosomes.
Polyploid
Organisms w/ 3 or more sets of chromosomes
Gene mutations
molecular changes in the DNA sequence of a gene
- can involve a base substitution
e.g. T -> G
Transition
a change of a pyrimidine (C. T) to another pyrimidine or a purine (A, G) to another purine
Transbersion
a change of a pyrimidine to a purine or vice versa
Silent mutations
base substitutions that don't alter the amino acid sequence of the polypeptide
Missense mutations
base substitutions in which an the amino acid change does occur
Nonsense mutations
base substitutions that change a normal codon to a stop codon
Frameshift mutations
involve the addition or deletion of a number of nucleotides that is not divisible by 3
shifts the reading frame so that translation of mRNA results in a completely different amino acid downstream of the mutation
Endopolyploidy
Diploid animals that sometimes produce polyploid tissues
Polytene Chromosomes
bundle of chromosomes that lie together in a parallel fashion
- bundle is made from doublings of drosophila chromosome pairs wherein chromosomes undergo repeated rounds of replication w/o cellular division
3 natural mechanisms that cause the
chromosome number to vary
1. Meiotic nondisjunction
2. Mitotic nondisjunction
3.
Interspecies crosses
Nondisjunction in meiosis I
failure of chromosomes to segregate properly during anaphase
During fertilization, some gametes produce an individual that is trisomic
Some gametes produce an individual that is monosomic
for the missing chromosome
All four gametes are abnormal
Nondisjunction in Meiosis II
If nondisjunction occurs in meiosis II,
50% Abnormal gametes
50% Normal gametes
Meiotic Nondisjunction
In rare cases, all the chromosomes can undergo nondisjunction and
migrate to one daughter cell
This is termed complete nondisjunction
- It results in one diploid cell and one without
chromosomes
- The cell without chromosome is inviable
- The
diploid cell can participate in fertilization with a
normal
gamete (this yields a triploid individual)
Mitotic Abnormalities
Abnormalities in chromosome number often occur after fertilization.
- if so, abnormality occurs during mitosis not meiosis
1. Mitotic nondisjunction: sister chromatids separate improperly (leads to trisomic and monosomic daughter cells)
2. Chromosome loss: one of sister chromatids do not migrate to a pole (leads to normal and monosomic cells)
Mosaicism
genetic abnormalities that occur after fertilization
- contains a subset of cells genetically different from the rest of the organism
- size and location of the mosaic region depends on the timing and location of the original abnormality
extreme case: abnormality takes place early embryonic development (precursor to be large segment of organism)
Comparison of Autopolyploidy, Alloploidy, and Allopolyploidy
The number of chromosome sets in a given individual or species can
vary in three ways
- Autopolyploidy: increase in the number of
sets within a single species
- Alloploidy: the combining of
chromosome sets from different species
- Allopolyploidy: when the
number of chromosome sets increases in an alloploid
Complete nondisjunction can produce an individual with one or
more sets of chromosomes
- diploid species -> autopolyploidy
(tetraploid)
A common mechanism for changes in the number of sets
of chromosomes is alloploidy
- the result of interspecies
crosses
- Most likely occurs between closely related species
- species 1, species 2 -> alloploidy (allodipoid)
An allodiploid has one set of chromosomes from two different
species.
An allopolyploid contains a combination of both
autopolyploidy and alloploidy.
An allotetraploid contains two
complete sets of chromosomes from two different species.
- species 1, species 2 -> alloploidy (allotetraploid)
