microbial genetics
what is a genome?
the sum total of genetic material of an organism, the most exists in the form of chromosomes, some may appear in non-chromosomal forms
what are the non-chromosomal forms of a genome?
plasmid and organelles
what is a plasmid?
tiny extra pieces of DNA
non-chromosomal genome in organelles?
mitochondria and chloroplasts have their own DNA
what is a chromosome?
a distinct cellular structure composed of a neatly packaged DNA molecule
what is a eukaryotic chromosome?
located in the nucleus; they vary in number from a few to hundreds; they can occur in pairs (diploid) or singles (haploid); they have a linear appearance
what is a bacterial chromosome?
usually single, circular (double-stranded) chromosome, although many bacteria have multiple, circular chromosomes and some have linear chromosomes
what is a gene?
basic informational packets in which a chromosome is subdivided containing the necessary code for a particular cell function
what are the three categories of genes?
structural genes that code for proteins
genes that code for the RNA machinery used in protein production
regulatory genes that control gene expression
what is a geneotype?
the sum of all types of genes constituting an organism's distinctive genetic makeup
what is a phenotype?
the expression of the genotype that creates certain structures or functions (traits)
structure of DNA
phosphate, deoxyribose sugar, nitrogenous base
nitrogenous bases
purines (A,G) and pyrimidines (T,C)
adenine always pairs with
thymine
guanine always pairs with
cytosine
DNA arrangement
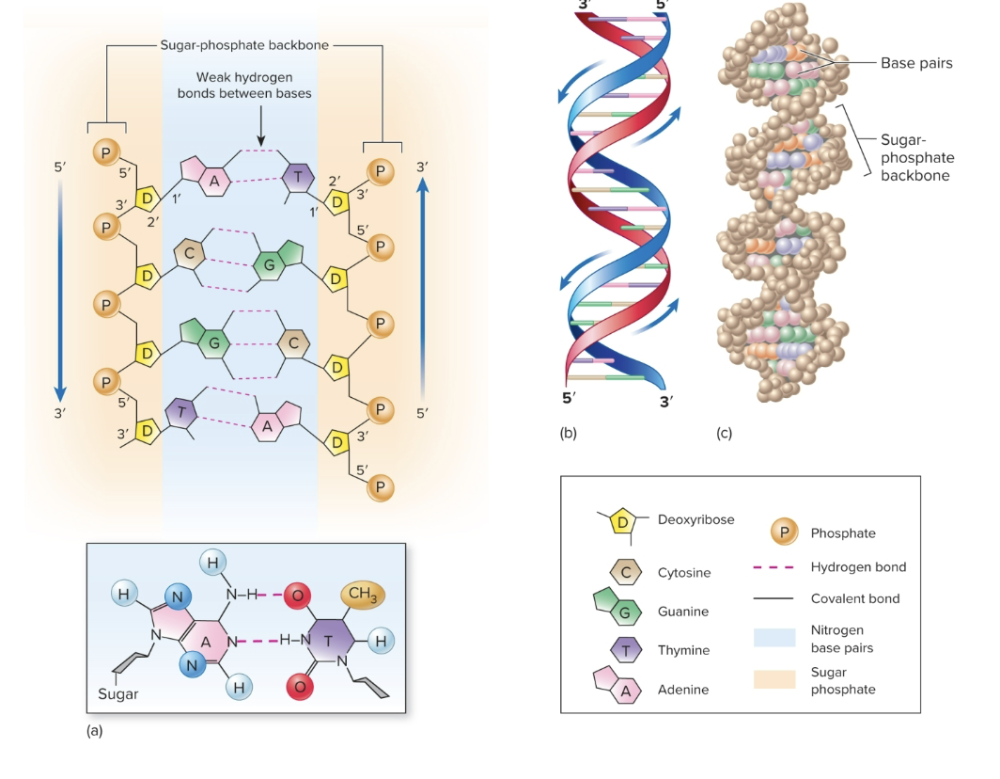
antiparallel
one side of the helix runs in the opposite direction of the other (5'-3' and 3'-5')
what are the steps of DNA replication?
semiconservative replication
each daughter molecule is identical to the parent in composition
neither is completely new
the template strand is an original parental DNA strand
helicase
unzipping the DNA helix
primase
synthesizing an RNA primer
DNA polymerase 3
adding bases to the new DNA chain and proofreading the chain for mistakes
functions once the DNA helix strands are unwound and separated
synthesizes a daughter strand of DNA using the parental strand as a template
can only add nucleotides to an existing chain 5'-3' direction
DNA polymerase 1
removing primer, closing gaps, and repairing mismatches
ligase
final binding of nicks in DNA during synthesis and repair
topoisomerase 1
making single-stranded DNA breaks to relieve supercoiling at origin
topoisomerase 2 and 4
making double-stranded DNA breaks to remove supercoiling ahead of origin and separate replicated daughter DNA molecules
replication fork
the place in the helix where the strands are unwound and replication is taking place
each circular DNA molecule will have two replication forks
primer
a length of RNA that is inserted initially during replication before being replaced by DNA
how does the synthesis of the leading strand differ from the synthesis of the lagging strand?
leading strand: new DNA synthesized continuously in a 5' to 3' direction
lagging strand: new strand must be synthesized in short segments (5' to 3'), later sealed together to form a strand in 3' to 5' direction
Okazaki fragments
short segments of DNA synthesized in a 5' to 3' direction which are then sealed together to form the 3' to 5' strand
the central dogma
transcription: DNA is used to synthesize RNA
translation: RNA used to produce proteins
why is the central dogma incomplete?
a wide variety of RNAs are used to regulate gene function
many genetic malfunctions that cause human disease are found in regulatory RNA, not in genes for proteins
the DNA that codes for these crucial RNA molecules was once called "junk" DNA
connection between DNA and an organism's traits
a protein's primary structure determines its characteristic shape and function
proteins ultimately determine phenotype
DNA mainly a blueprint that tells the cell which kinds of proteins to make and how to make them
participants in transcription and translation
mRNA
tRNA
rRNA
ribosomes
several types of enzymes
many raw materials
mRNA
transports the DNA master code to the ribosome
synthesized in a process similar to synthesis of the leading strand during DNA replication
tRNA
brings amino acids to ribosome during translation
rRNA
forms the major part of a ribosome and participates in protein synthesis
3-dimensional shapes that create the two subunits of the ribosome
miRNA
regulation of gene expression and coiling of chromatin
primer
primes DNA
ribozymes and spliceosomes
remove introns from other RNAs in eukaryotes
codon
a series of triplet bases that hold the message of the transcribed mRNA
anticodon
found at the bottom loop of the cloverleaf
designates the specificity of the tRNA and complements the mRNA codon
redudancy
certain amino acids are represented by multiple codons
allows for the insertion of correct amino acids even when mistakes occur in the DNA sequence
wobble
only the first two nucleotides are required to encode the correct amino acid
the third nucleotide does not change its sense
permits some variation or mutation without altering the message
start codon
the first 3 RNA nucleotides that signal the beginning of the message
AUG
stop codons
nonsense codons - one of three codons that has no corresponding tRNA and causes translation to be terminated
UAA, UAG, UGA
introns
intervening sequences of bases that do not code for protein
excised into lariats by spliceosome
exons
coding regions
promoter sequence
recognized by RNA polymerase
operator
acts as an on/off switch for transcription
regulator
composed of the gene that codes for the repressor, a protein capable of repressing the operon
A site
A site accepts incoming tRNA molecules carrying amino acids
P site
P site holds the tRNA attached to the growing polypeptide chain until the next amino acid is added
operons
coordinated set of genes regulated as a single unit
found only in bacteria and archaea
can be inducible or repressible, determined by how transcription is affected by the environment surrounding the cell
transcription factors
insert into the grooves of the DNA molecule and enhance the transcription of specific genes
regulate gene expression in response to environmental stimuli
DNA "knot"
cytosines bind to other cytosines forming a "knot" in the helix of DNA
blocks the promoter region of genes in order to stop transcription
drugs that inhibit protein synthesis
rifamycins
actinomycin D
drugs that interfere with the ribosome
erythromycin
spectinomycin
recombination
an event in which one bacterium donates DNA to another bacterium
recombinant
any organism that contains and expresses genes that originated in another organism
horizontal gene transfer
any transfer of DNA that results in organisms acquiring new genes that did not come directly from parent organisms
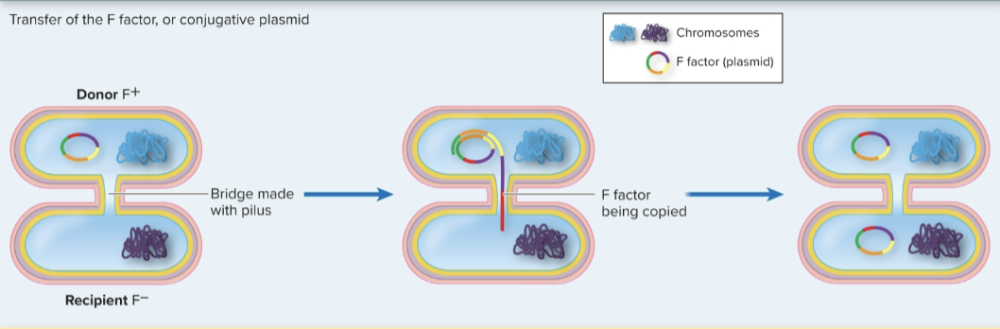
conjugation
direct
donor cell with pilus
fertility plasmid in donor, both donor and recipient alive, bridge forms between cells to transfer DNA
transformation
indirect
free donor DNA (fragment), live; competent recipient cell
the acceptance by a bacterial cell of small fragments of soluble DNA from the surrounding environment
transduction
indirect
toxins; enzymes for sugar fermentation; drug resistance
conjugation F factor
resistance (R) plasmids or factors
carry genes for resisting antibiotics or other drugs
commonly shared among bacteria through conjugation
can confer multiple resistance to antibiotics
R factors can also carry genetic codes for resistance to heavy metals or for synthesizing virulence factors
competent
cells that are capable of accepting genetic material through transformation
generalized transduction
random fragments of disintegrating host DNA are taken up by a phage during assembly
any gene from the bacterium can be transmitted
specialized transduction
a highly specific part of the host genome is incorporated into the virus when the prophage DNA separates from the chromosome (carrying host genes with it)
transposable elements
jumping genes shift from one part of the genome to another
from one chromosomal site to another, from a chromosome to a plasmid, and from a plasmid to a chromosome
insertion elements
the smallest TEs consist only of two tandem repeats
retroptransposon
a type of TE that can transcribe DNA into RNA and then back into DNA for insertion in a new location
general effects of TEs
scramble genetic language
can be beneficial or adverse, depending on: where the insertion occurs in a chromosome, what kind of genes are relocated, the type of cell involved
effects of TEs in bacteria
changes in colony morphology, pigmentation, and antigenic characteristics
replacement of damaged DNA
transfer of drug resistance between bacteria
pathogenicity islands
contain multiple genes that are coordinated to create a new trait in bacterium, making it pathogenic
mutation
any change to the nucleotide sequence in the genome
the driving force of evolution
in microorganisms, mutations become evident in altered gene expression, such as altered pigment production or development of resistance to a drug
wild type mutation
a microorganism that exhibits a natural, non-mutated characteristic
the trait present in the highest numbers in a population
mutant strain
an organism that bears a mutation
spontaneous mutation
a random change in the DNA arising from errors in replication that occur randomly
induced mutations
result from exposure to known physical or chemical agents that damage DNA (known as mutagens)
nitrous acid, bisulfite
chemical agent
remove an amino group from some bases
ethidium bromide
chemical agent
inserts between the paired bases
acridine dyes
chemical agent
cause frameshift due to insertion between base pairs
nitrogen base analogs
chemical agent
compete with natural bases for sites of replicating DNA
ionizing (gamma rays, X rays)
radiation agent
form free radicals that cause single or double breaks in DNA
ultraviolet
radiation agent
causes cross-links between adjacent pyrimidines
point mutation
small mutation that affects only a single base on a gene
involve addition, deletion, or substitution of single bases
lethal mutation
mutation that leads to cell dysfunction or death
neutral mutation
produce neither adverse nor helpful changes
missense mutation
any change in the code that leads to the placement of different amino acids
nonsense mutation
changes in normal codon into a stop codon
silent mutation
alters a base but does not change the amino acid
the redudancy by a change in the third base of the codon
back mutation
occurs when a gene that has undergone mutation reverses to its original base composition
frameshift mutation
occurs when one or more bases are inserted into or deleted from a newly synthesized DNA strand
alters the reading frame of mRNA
nearly always results in a nonfunctional protein
photoactivation
light repair of damage caused by ultraviolet radiation
requires visible light and a light-sensitive enzyme called DNA photolyase, which can detect and attach to the damaged areas
mismatch repair
a repair system can locate mismatched bases that were missed during proofreading
the base must be replaced soon after the mismatch is made, or the repair enzymes will not recognize it
excision repair
mutations are excised by a series of enzymes that remove the incorrect bases and add the correct ones
the ames test
used to rapidly detect chemicals with carcinogenic potential
uses bacteria (salmonella typhimurim) rather than experimental animals
histone + colonies arising from spontaneous back-mutation on control plate
histone + colonies in presence of chemical on test plate