BMD 315 Module 1 Study Guide/ Learning Objectives
What is the structure of an atom?
An atom is the basic unit of matter and is made up of three main parts:
Protons positively charged particles found in the nucleus.
Neutrons neutral particles also in the nucleus.
Electrons negatively charged particles that orbit the nucleus in electron shells or energy levels.
What is the structure of an ion?
An ion is an atom or molecule that has gained or lost electrons, giving it a net charge: Cation: positive ion (lost electrons) Anion: negative ion (gained electrons)
Ionic bond
Formed when electrons are transferred from one atom to another.
Typically between a metal and a non-metal. Results in oppositely charged ions that attract each other.
Covalent bond
Formed when atoms share electrons.
Usually occurs between non-metal atoms. Can involve single, double, or triple electron pairs.
Hydrogen bond
A weak attraction between a hydrogen atom (already covalently bonded to something electronegative like oxygen or nitrogen) and another electronegative atom. Important in stabilizing biological structures like DNA and proteins.
Nonpolar covalent bonds
Electrons are shared equally between atoms.
Happens when atoms have similar or identical electronegativities (ability to attract electrons).
There is no partial charge on the atoms.
Key Feature:
Molecule has no poles (no distinct positive or negative ends).
Polar covalent bonds
Electrons are shared unequally.
One atom attracts electrons more strongly (is more electronegative), creating:
a slightly negative (6-) end (where electrons hang out more) and a slightly positive (5*) end.
Ex: In water, oxygen is more electronegative than hydrogen, so electrons spend more time near oxygen.
Key Feature:
Molecule has positive and negative poles, like a magnet.
Why Are lons and Polar Molecules Soluble in Water?
Water is a polar molecule: It has a partial negative charge near oxygen and a partial positive charge near the hydrogen atoms.
Polar molecules and ions are attracted to the opposite charges on water molecules. This attraction helps break them apart (dissolve them) in water.
Acid
A substance that releases H + (hydrogen ions) in solution.
Increases the concentration of hydrogen ions in a solution.
Acidic/ Basic
A solution is acidic if it has more H+ ions than OH/ pH is less than 7.
A solution is basic if it has more OH- ions than H+/ pH is greater than 7.
Base
A substance that removes H+ ( hydrogen ions) from a solution or releases OH- (hydroxide ions).
Decreases the concentration of hydrogen ions.
What is pH and how does it relate to the concentration of H+?
pH is a scale that measures how acidic or basic a solution is.
It is based on the concentration of hydrogen ions (H+).
Formula: pH -log[H+]
So, as [H+] increases, pH decreases (more acidic).
As [H+] decreases, pH increases (more basic).
What is a buffer?
a system that resists changes in pH when small amounts of acid or base are added. It usually consists of a weak acid and its conjugate base.
In your body, a key buffer is the bicarbonate buffer system, important for maintaining blood pH around 7.35-7.45.
The Bicarbonate buffer pair
In your body:
Carbonic acid (H2CO3-)= weak acid
Bicarbonate ion (HCO3-) = weak base
They work together to keep your blood from getting too acidic or too basic.
If the blood becomes too acidic:
The bicarbonate ion (HCO3-) picks up the extra H+. Then carbonic acid breaks down into water and carbon dioxide, which is breathed out.
If the blood becomes too basic (too much OH-): The carbonic acid gives up a hydrogen ion to neutralize the OH-.
How Carbon bonds with other atoms and itself?
(Via Covalent bonds, max = 4 bonds)
Other Carbons- Forms chains, rings, or branches. Can make single (-), double (=), or triple (=) bonds
Hydrogen (H)- Forms simple C-H bonds Found in fuels like methane (CH4)
Oxygen (O)- Can form: C-0 (single bond) or C=0 (double bond)
Nitrogen (N)- Can form: C-N (single bond) or C=N (triple bond)
Different types of Carbohydrates
Monosaccharides (simple sugars)
One sugar unit, Quick energy source
Examples: Glucose, Fructose, and Galactose
Disaccharides (two sugar units)
Ex: Sucrose = glucose + fructose, Lactose = glucose + galactose, and Maltose= glucose + glucose
Polysaccharides (many sugar units)
Long chains of sugars that are used for energy storage or structure
Examples: Starch, Glycogen, and Cellulose
Different types of lipids
Fats and Oils (Triglycerides)
Made of 1 glycerol + 3 fatty acids
Saturated Fats [Fats solid at room temp (animal fat)]
Unsaturated Fats [Oils = Iiquid at room temp (olive oil)]
Phospholipids
Main part of cell membranes
Have a water-loving head and a water-hating tail
Example: Phospholipids in your cell walls
Steroids
Ring-shaped lipids that are used as hormones or cholesterol
Examples:
Cholesterol (in cell membranes)
Testosterone and estrogen
Dehydration synthesis (build up) of Carbohydrates and Lipids
In Monosaccharides (simple sugars like glucose) join to form disaccharides or polysaccharides.
Example: Glucose + Glucose -> Maltose + Water What happens: An -OH (hydroxyl) group from one sugar and an H from another sugar are removed. They form H2O (water). The sugars bond via a glycosidic bond.
In Triglycerides (fats): Made by joining 1 glycerol and 3 fatty acids.
Example: Glycerol +3 Fatty Acids -> Triglyceride + 3 Water
What happens: Each fatty acid attaches to glycerol by removing a water molecule. The bond formed is called an ester bond.
Hydrolysis (break down) of Carbohydrates and Lipids
In Carbohydrates:
Breaks polysaccharides into monosaccharides.
Example: Maltose + Water -> Glucose + Glucose
What happens: A water molecule is added. The glycosidic bond is broken.
In Triglycerides:
Breaks fats into glycerol + 3 fatty acids.
Example:
Triglyceride + 3 Water -> Glycerol + 3 Fatty Acids
What happens:
Water helps break the ester bonds.
Describe the nature of Phospholipids
Found in cell membranes.
Made of: 2 fatty acid tails (hydrophobic – "water-fearing"), 1 phosphate group head (hydrophilic – "water-loving") and 1 glycerol backbone
Nature:
Amphipathic: Has both hydrophobic and hydrophilic parts.
In water, they form bilayers, which are the basis of cell membranes.
The hydrophobic tails face inward, away from water, while hydrophilic heads face outward, toward water.
Describe the nature of Prostaglandins
Derived from fatty acids
Not part of membranes—these act more like hormone-like messengers.
Have a five-membered carbon ring and two side chains.
Hydrophobic but work in watery environments
Nature: Act locally, not throughout the body (unlike hormones).
- Involved in: Inflammation, Pain, Fever, Blood clotting, Smooth muscle contraction (e.g., in uterus and lungs)
Description of amino acids
the building blocks of proteins.
Each amino acid has the same basic structure: Amino group (—NH₂), Carboxyl group (—COOH), Hydrogen atom (—H), and R group (side chain) – varies between amino acids and determines their properties.
All four parts are bonded to a central carbon (called the alpha carbon).
There are 20 common amino acids, each with a unique R group.
How peptide bonds are formed?
A peptide bond links amino acids together to form proteins or polypeptides.
Amino acid 1's —COOH (carboxyl group) + Amino acid 2's —NH₂
(amino group)
↓ ↓
(remove H₂O → dehydration
synthesis)
↓
Result: —CO—NH— (peptide bond)
How are peptide bonds broken down?
To break a peptide bond, water is added (hydrolysis)
The bond is broken, restoring the original —COOH (carboxyl group) and —NH₂ groups (amino group) on the separate amino acids.
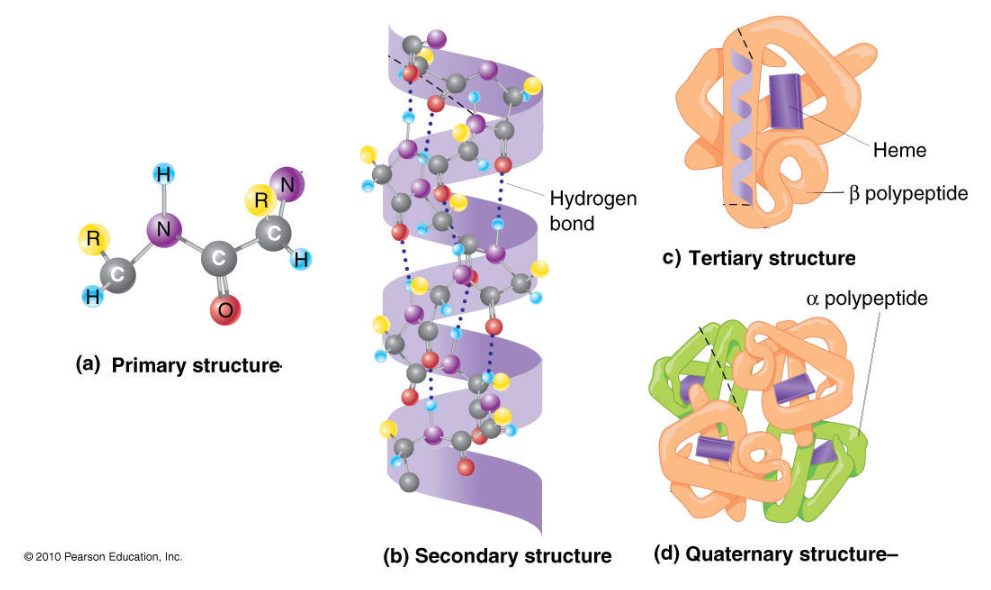
The Four Orders of protein structures
Primary Structure
The sequence of amino acids in a polypeptide chain. It is held by Peptide bonds.
- Importance: Like letters in a sentence — the exact order matters for the final function.
Secondary Structure
Local folding of the chain into α-helices (coils) and β-pleated sheets (folds) . It is held by Hydrogen bonds between nearby backbone atoms.
- Importance: Provides flexibility and shape.
Tertiary Structure
The overall 3D shape of a single polypeptide. It is held by: Hydrogen bonds, Ionic bonds, Disulfide bridges, Hydrophobic interactions
- Importance: Determines the protein’s functional shape.
Quaternary Structure
- Definition: When two or more polypeptide chains come together to form one protein. It is held by: Same interactions as tertiary.
- Examples: Hemoglobin (has 4 polypeptide chains)
The different functions of proteins
Enzymes: Speed up chemical reactions. Ex: Amylase, DNA polymerase
Transport: Carry substances across membranes or through blood. Ex: Hemoglobin, ion channels
Structural: Provide support and shape to cells and tissues. Ex: Collagen, keratin Defense: Fight infection. Ex: Antibodies (immunoglobulins)
Signaling: Carry messages between cells. Ex: Insulin, growth hormone
Movement: Help cells or muscles move. Ex: Actin, myosin
Storage: Store ions or amino acids. Ex: Ferritin (iron storage)
How Structure Gives Function Specificity?
A protein's shape = function.
The 3D structure of a protein allows it to fit like a lock and key with its specific target (like a substrate for an enzyme).
Even a small change in the amino acid sequence (primary structure) can change the folding, shape, and destroy or alter the function.
Ex: Enzyme active sites are shaped to bind only to specific molecules.
- If the structure is damaged (by heat, pH change, or mutation), it becomes denatured and no longer works.
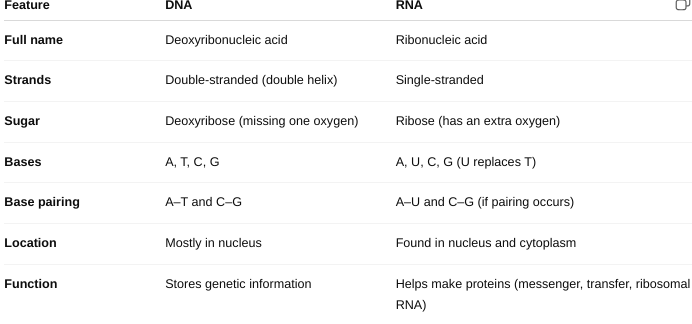
Describe the structure of a nucleotide
the basic building block of nucleic acids
(DNA and RNA).
Each nucleotide
has three parts:
Phosphate group (PO₄³⁻)
Sugar
- DNA: Deoxyribose
- RNA: Ribose
Nitrogenous base
- Purines: Adenine (A) and Guanine (G)
- Pyrimidines: DNA: Cytosine (C) and Thymine (T) or RNA: Cytosine (C) and Uracil (U)
The structural differences between DNA and RNA
What is the law of complementary base pairing?
states that in DNA, specific nitrogenous bases always pair with each other in a consistent way:
Adenine (A) always pairs with Thymine (T)
Cytosine (C) always pairs with Guanine (G)
How does the law of complementary base pairing work between DNA strands?
DNA is made of two strands twisted into a double helix. These strands are held together by hydrogen bonds between the bases.
A–T form 2 hydrogen bonds
C–G form 3 hydrogen bonds
Because of this rule:
One strand determines the sequence of the other.
For
example: Strand 1: A - T - G - C
Strand 2: T - A - C - G
It ensures accurate DNA replication—each strand serves as a template, maintains genetic stability over generations and prevents random or mismatched pairing, which would lead to mutations.
Describe the structure of the plasma (cell) membrane
Function: Acts as a selective barrier, controlling what enters and leaves the cell.
Structure: Made of a phospholipid bilayer
Hydrophilic heads (face water) and Hydrophobic tails (face inward)
Contains: Proteins (for transport, signaling, and structure), Cholesterol (adds stability and fluidity), and Carbohydrates (for cell recognition and communication)
Key Feature:
- Fluid Mosaic Model: Membrane is flexible and proteins float like "tiles" in a fluid lipid "sea."
Describe the structure of cilia
Function: Move fluids across the surface of cells (e.g., mucus in the respiratory tract) and can also play sensory roles.
Structure: Short, hair-like projections, made of microtubules in a 9 + 2 arrangement:
9 pairs of microtubules around the outside and 2 single microtubules in the center
- Anchored to the cell by a basal body (like a foundation)
Describe the structure of flagella
Function: Move the entire cell (like sperm cells swimming)
Structure: Long, tail-like projection, the same 9 + 2 microtubule structure as cilia and also anchored by a basal body
Amoeboid Movement
A type of movement used by cells like amoebas and white blood cells.
The cell forms pseudopods (“false feet”) by extending part of its cytoplasm. The cytoplasm flows into the pseudopod, pulling the cell forward. This is driven by the cytoskeleton (especially actin filaments).
Function: Allows crawling movement and helps cells hunt or move through tissues.
Phagocytosis ("Cell Eating")
A form of endocytosis where the cell engulfs large particles (e.g., bacteria, dead cells).
The membrane wraps around the target and forms a vesicle called a phagosome. The phagosome then fuses with a lysosome, which digests the material.
Function : Used by immune cells like macrophages to destroy invaders
Pinocytosis ("Cell Drinking")
Another form of endocytosis where the cell takes in fluids and small molecules. The membrane pinches inward, forming a small vesicle of fluid.
Function: Helps cells take in nutrients and maintain fluid balance.
Receptor-Mediated Endocytosis
A selective form of endocytosis.
Receptors on the cell surface bind to specific molecules (like cholesterol or hormones). Once bound, the membrane folds inward to form a vesicle.
Function: Allows cells to take in specific substances in concentrated amounts.
Exocytosis
Opposite of endocytosis.
A vesicle inside the cell fuses with the plasma membrane, releasing its contents outside the cell. Used to remove waste or release hormones, enzymes, or neurotransmitters.
Function: Secretes important substances and maintains membrane balance.
The structure and functions of cytoskeleton
Structure: A network of protein fibers inside the cell.
Made of:
Microfilaments (actin): thin, flexible fibers
Intermediate filaments: medium-sized, rope-like
Microtubules: thick, hollow tubes (made of tubulin)
Functions: Supports cell shape, helps with movement (inside and outside the cell), guides organelle positioning and transport, and forms cilia and flagella
The structure and functions of lysosomes
Structure: Small, membrane-bound vesicles filled with digestive enzymes
Functions: Break down worn-out cell parts, bacteria, and macromolecules, known as the cell’s "recycling center" and important for immune responses and cell cleanup
The structure and functions of peroxisomes
Structure: Small, membrane-bound organelles (like lysosomes) and contain enzymes (like catalase) that break down toxins
Functions: Break down fatty acids, detoxify harmful substances (e.g., hydrogen peroxide into water and oxygen), and protect cells from oxidative damage
The structure and functions of mitochondria
Structure: Bean-shaped organelles with double membranes
Outer membrane: smooth
Inner membrane: folded into cristae for surface area
Inside: matrix (fluid-filled space)
Functions: Produce ATP (cell’s energy) through cellular respiration, known as the “powerhouse” of the cell and also involved in apoptosis (controlled cell death)
The structure and functions of ribosomes
Structure: Small, round structures made of RNA and proteins
- Found: Free in cytoplasm,and attached to rough ER
Functions: Make proteins by linking amino acids together using instructions from mRNA
- Free ribosomes → proteins for use inside the cell
- ER-bound ribosomes → proteins for membranes or export
The structures and functions of the endoplasmic reticulum
Rough ER (RER)
Structure: A network of flattened membranes with ribosomes attached to the surface.
Function: Makes proteins (with the help of ribosomes), modifies proteins (e.g., adds sugar chains), and sends proteins to the Golgi in transport vesicles
Smooth ER (SER)
Structure: Similar to rough ER, but no ribosomes and more tubular in shape
Function: Makes lipids (fats, oils, steroid hormones), detoxifies drugs and alcohol, and stores calcium in muscle cells
The structure and functions of the Golgi Complex (Golgi Apparatus)
Structure: A stack of flattened, membrane-bound sacs (like pancakes)
Function: Receives proteins and lipids from the ER, modifies them (e.g., adds sugar or phosphate groups), sorts and packages them into vesicles, and send them to their final destinations (inside or outside the cell)
How do the ER and Golgi Apparatus work together?
Proteins/lipids are made in the ER
- Proteins in rough ER and Lipids in smooth ER
Transport vesicles carry them from the ER to the Golgi
- In the Golgi: Molecules are modified, sorted, and repackaged
- The Golgi sends finished products: To the plasma membrane (for secretion or membrane use), to lysosomes, to other parts of the cell
The structure of the nucleus
Function: Controls the cell's activities and stores genetic information (DNA).
Structure:
Nuclear envelope: Double membrane that surrounds the nucleus; has pores to let things in/out.
Nucleoplasm: Jelly-like fluid inside the nucleus.
Nucleolus: Dense area that makes ribosomes.
Chromatin: DNA + proteins
The structure of chromatin
Chromatin = DNA + histone proteins
- Found inside the nucleus. DNA wraps around histones to stay organized and compact.
- Chromatin exists in two forms:
- Euchromatin: Loosely packed, light-staining, an Active DNA (being used for RNA)
- Heterochromatin: Densely packed, dark-staining an Inactive DNA (not being used)
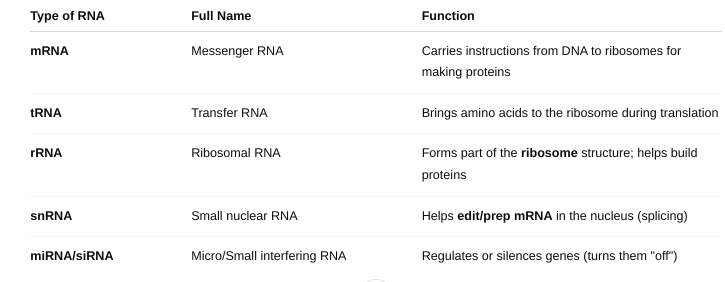
The types of RNA
How DNA directs the synthesis of RNA in genetic transcription?
Transcription is the process by which a gene
on DNA is copied into RNA.
It’s the first
step in making a protein (DNA → RNA → Protein).
Steps of Transcription
Initiation: The enzyme RNA polymerase binds to a promoter (a specific DNA sequence at the start of a gene). This signals where transcription should start.
Elongation: RNA polymerase unzips the DNA strand (just one section).
- It reads one strand of DNA (called the template strand) and
builds a complementary RNA strand.
- DNA A → RNA U
- DNA T → RNA A
- DNA C → RNA G
- DNA G → RNA C
- RNA is built in the 5′ to 3′ direction.
Termination: When RNA polymerase reaches a termination sequence, it stops. The new RNA strand (usually mRNA) detaches.The DNA zips back up.
The Key Players in Transcription
- DNA template- Holds the gene code
- RNA polymerase- Enzyme that builds RNA
- Promoter- Start signal for transcription
- Terminator- Stop signal for transcription
- mRNA- Messenger RNA copy that leaves the nucleus
How RNA directs the synthesis of proteins in genetic translation?
Translation is the process where the genetic code in mRNA is used to build a protein (a chain of amino acids).
It’s the second step of gene expression:
DNA → RNA → Protein
mRNA is read in groups of 3 bases called codons. Each codon codes for one amino acid.
Steps of Translation
Initiation: The ribosome attaches to the start codon (AUG) on the mRNA. The first tRNA brings the amino acid methionine.
Elongation: The ribosome reads the next codon on the mRNA. A tRNA with the matching anticodon brings the correct amino acid. The ribosome links amino acids together with peptide bonds to form a chain.
Termination: When the ribosome reaches a stop codon (e.g., UAA, UAG, UGA), translation stops. The protein chain is released and folded into its final shape.
The Key Players in Translation

How proteins are modified after translation?
After a protein is made through translation, it often needs to be modified to become functional. These changes are called post-translational modifications.
Folding: Protein folds into its correct 3D shape (often with help from chaperone proteins)
Cleavage: A piece of the protein is cut off to activate it
Phosphorylation: A phosphate group is added (can turn protein on/off)
Glycosylation: Sugar chains are added (important for cell recognition and signaling)
Lipidation: Lipid added for membrane anchoring
Disulfide bonds: Help stabilize protein structure
These modifications happen in the endoplasmic reticulum (ER), Golgi apparatus, or cytoplasm depending on the protein’s role.
How ubiquitin and the proteasome are involved in breaking down proteins?
Sometimes proteins are: Misfolded, Damaged and No longer needed
They must be removed to keep the cell healthy.
Ubiquitin: A small protein that acts like a "tag".
It is attached to proteins that need to be destroyed. The process is called ubiquitination. Multiple ubiquitin tags = “this protein must go!”
Proteasome: A large protein complex (like a molecular paper shredder).
It recognizes and destroys proteins that are tagged with ubiquitin. It breaks them into small peptides or amino acids that the cell can reuse.
Explain the semiconservative replication of DNA in DNA synthesis.
Semiconservative replication means that when DNA is copied, each new DNA molecule contains one original (parent) strand and one new strand.
Steps of DNA Synthesis (Replication)
Unwinding the DNA
The enzyme helicase unzips the double helix by breaking the hydrogen bonds between base pairs. This forms a replication fork (Y-shaped opening).
Priming
Primase lays down a short RNA primer to start the new strand.
Building New Strands
DNA polymerase adds new nucleotides to the exposed bases using base pairing rules:
A pairs with T, C pairs with G, One strand (the leading strand) is built continuously. The other strand (the lagging strand) is built in pieces called Okazaki fragments and later joined by DNA ligase.
Result
Two identical DNA molecules are formed.
- Each has: 1 original strand (from the parent molecule) and 1 newly synthesized strand
The Cell Cycle
the series of stages a cell goes through to grow and divide into two identical daughter cells.
Phases of the Cell Cycle
Interphase – the longest phase (cell prepares to divide)
- G1 phase ("Gap 1") - Cell grows, performs normal functions
- S phase ("Synthesis")- DNA is replicated
- G2 phase ("Gap 2")- Cell continues to grow; checks for errors
Mitotic Phase (M phase) – actual cell division
- Mitosis: Nucleus divides (has 4–5 sub-phases: prophase, metaphase, anaphase, telophase)
- Cytokinesis: Cytoplasm divides, forming 2 new cells
G0 phase – some cells exit the cycle permanently (e.g., nerve cells)
Factors that affect the cell cycle
- Growth factors: Proteins that signal the cell to divide
- Nutrients & energy: Cells need fuel and materials to grow
- Cell size: Too small = keep growing; right size = divide
- DNA damage: Stops the cycle for repair or leads to apoptosis
- Checkpoints: Built-in stops at G1, G2, and M to ensure accuracy
- Hormones: Can stimulate or block cell division
If any errors or damage are detected, the cell can pause the cycle or self-destruct.
The significance of apoptosis
A controlled, safe way for cells to self-destruct when they’re old, damaged, or no longer needed.
Protects the body by removing damaged or cancer-prone cells, Shapes the body during development (e.g., removing webbing between fingers) and Maintains balance between cell growth and death
Failure in apoptosis can lead to: Cancer (if damaged cells survive and divide), Autoimmune diseases (if immune cells that should die survive), and Degenerative diseases (if too many cells die)
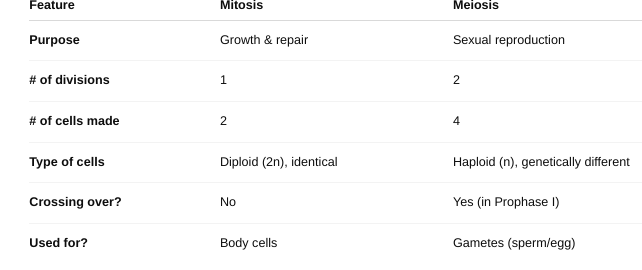
The Phases of Mitosis
Purpose: To create two identical diploid
cells (same number of chromosomes as the original
cell).
Used in: Body (somatic) cells.
Prophase: Chromosomes condense and become visible, Spindle fibers form, and Nuclear envelope breaks down
Metaphase: Chromosomes line up in the middle of the cell
Anaphase: Sister chromatids are pulled apart to opposite sides
Telophase: Chromosomes uncoil, and Nuclear envelopes reform around each set
Cytokinesis (not part of mitosis but follows it)- Cytoplasm divides, forming two identical daughter cells
The Phases of Meiosis
Purpose: To create four genetically unique
haploid cells (with half the chromosome number).
Used
in: Gametes (sperm and egg cells).
Meiosis I – Separates homologous chromosomes
Prophase I: Chromosomes pair up (synapsis), Crossing over occurs (genes are exchanged)
Metaphase I: Paired chromosomes line up in the middle
Anaphase I: Homologous chromosomes separate (not sister chromatids)
Telophase I & Cytokinesis: Two haploid cells form (each with half the chromosomes)
Meiosis II – Separates sister chromatids (like mitosis)
Prophase II: New spindle fibers form in each haploid cell
Metaphase II: Chromosomes line up in the center
Anaphase II: Sister chromatids are pulled apart
Telophase II & Cytokinesis: Four unique haploid cells are formed
Meiosis vs. Mitosis