Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
Microbiology Exam 3 Review - Chapter 6 + 21
front 1 Chapter 6 | back 1 Microbial Genetics |
front 2 What is a genome? | back 2 The sum total of genetic material of an organism
|
front 3 What are some nonchromosomal forms of genome? | back 3 1. Plasmids, which are tiny extra pieces of DNA, non-essential to the survival of the organism 2. Organelle: mitochondria and chloroplasts have their own DNA |
front 4 Genomes of cells are made of DNA, but viruses can contain ____ or _____. | back 4 DNA or RNA |
front 5 What is a chromosome? | back 5 A distinct cellular structure composed of a neatly packaged DNA molecule |
front 6 Where can genomes be found in eukaryotes? | back 6 Plasmids, Chloroplast DNA, Mitochondrial DNA, and Chromosomes
|
front 7 Where can genomes be found in bacteria? | back 7 Chromosomes (no nucleus) and plasmids
|
front 8 Where can genomes be found in viruses? | back 8 DNA and RNA in the capsid |
front 9 What are genes? | back 9 Basic informational packets in which a chromosome is subdivided containing the necessary code for a particular cell function |
front 10 What is the difference between genome, chromosomes, and genes? | back 10
|
front 11 Genes can be used for what three functions: | back 11
|
front 12 What is the difference between genotype and phenotype? | back 12 Genotype is the sum of all types of genes constituting an organism's distinctive genetic makeup. Phenotype is the expression of the genotype that creates certain structures or functions. |
front 13 True or False:
| back 13 TRUE Step 1: What this means
So, the statement is saying: Every organism carries a full set of genetic instructions (genotype), but only a portion of those genes are actively expressed to produce the observable traits (phenotype). |
front 14 What is the basic unit of a DNA molecule? | back 14 A nucleotide:
|
front 15 What are the four nitrogenous bases and what do they pair with? | back 15
Purines: A and G (Pure as Silver)
Pyrimidines: CUT the Pyramid
|
front 16 True or False: The typical bacteria chromosomes contains a few nucleotides. | back 16 FALSE: several million |
front 17 What are the differences between DNA and RNA? | back 17 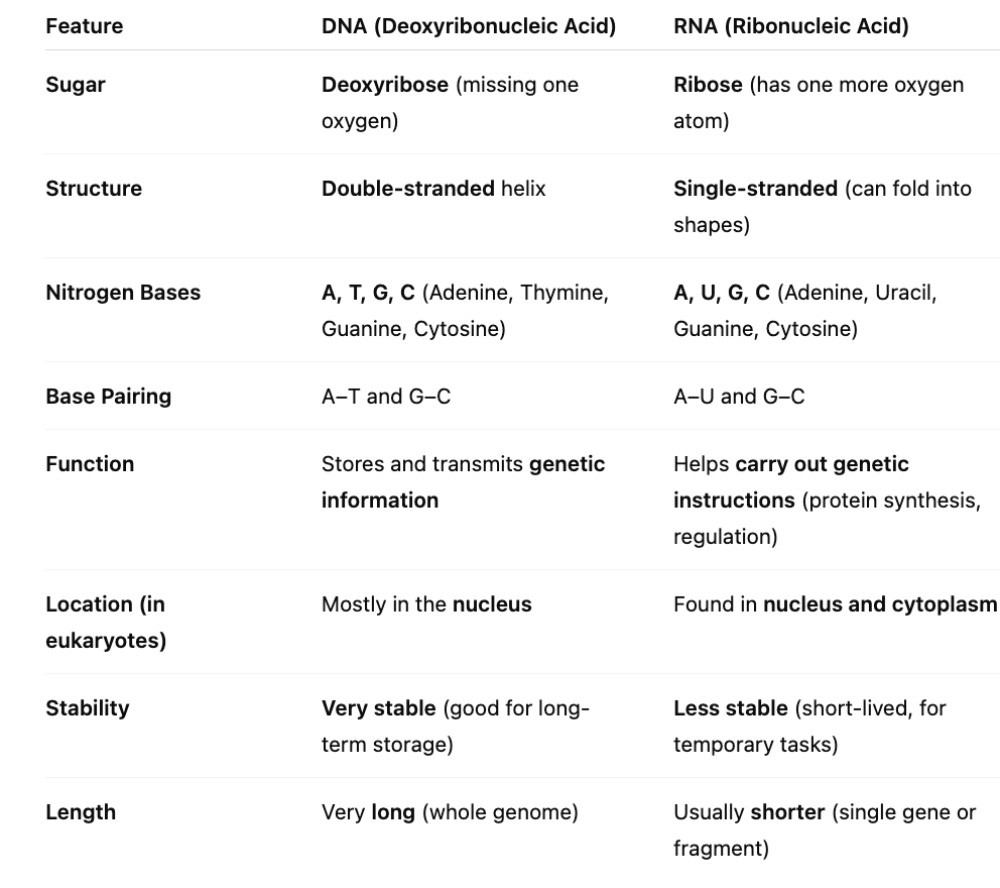 DNA also runs in an antiparallel arrangement (one side of the helix runs in the opposite direction of the other)
This is important in DNA synthesis (replication) and protein production RNA
|
front 18 Each strand provides a _______ for the replication of a new molecule guaranteeing accurate duplication | back 18 template |
front 19 True or False: The precise sequence of bases constitutes a genetic program responsible for the unique qualities of each organism | back 19 True |
front 20 What is the difference in replication forks and sites for prokaryotes vs. eukaryotes? What is the similarity between both? | back 20
Eukaryotic DNA is larger and has multiple origins of replication, allowing replication to occur at several sites simultaneously. Prokaryotic DNA, which is circular, begins replication at a single origin called OriC. Despite these differences, both eukaryotic and prokaryotic DNA replication are semi-conservative, meaning each new DNA molecule contains one original strand and one newly synthesized strand. |
front 21 Why do we study prokaryotic models compared to eukaryotic models? | back 21
In short, prokaryotes are simpler and more accessible models that reveal the same fundamental mechanisms used in eukaryotic DNA replication. |
front 22 Enzymes involved in prokaryote replication: Helicase | back 22 Unwinds the DNA by breaking the hydrogen bonds holding the two strands together, resulting in two seperate strands. |
front 23 Enzymes involved in prokaryote replication: Primase | back 23 Synthesizes and lays down an RNA primer to add nucleotides which allow the nucleotides to continue replication.
|
front 24 Enzymes involved in prokaryote replication: DNA Polymerase III | back 24 Adding bases to the new DNA chain and proofreading the chain for mistakes. Lays down DNA nucleotides for the new strand.
|
front 25 Enzymes involved in prokaryote replication: DNA Polymerase I | back 25 Removes the primer laid by primase, closes gaps, and repairs any mismatches |
front 26 Enzymes involved in prokaryote replication: Ligase | back 26 Final binding of the nicks in DNA during synthesis and repair Seals the primers and the strands together (especially in the lagging strand) |
front 27 Enzymes involved in prokaryote replication: Topoisomerase 1 | back 27 Make single-stranded DNA breaks to relieve supercoiling at the origin Helicase unwinds the DNA strands. |
front 28 Enzymes involved in prokaryote replication: Topoisomerase 2 (DNA gyrase) and 4 | back 28 Makes double-stranded DNA breaks to remove supercoiling ahead of origin and separate replicated daughter DNA molecules |
front 29 What is the overall replication process? It is known as ____________. | back 29
|
front 30 The template strand is an ________ parental DNA strand. | back 30 original |
front 31 In DNA replication, what is the leading strand? | back 31
|
front 32 In DNA replication, what is the lagging strand? | back 32
|
front 33 In DNA replication, what are Okazaki fragments? | back 33
|
front 34 Picture associated with DNA replication | back 34 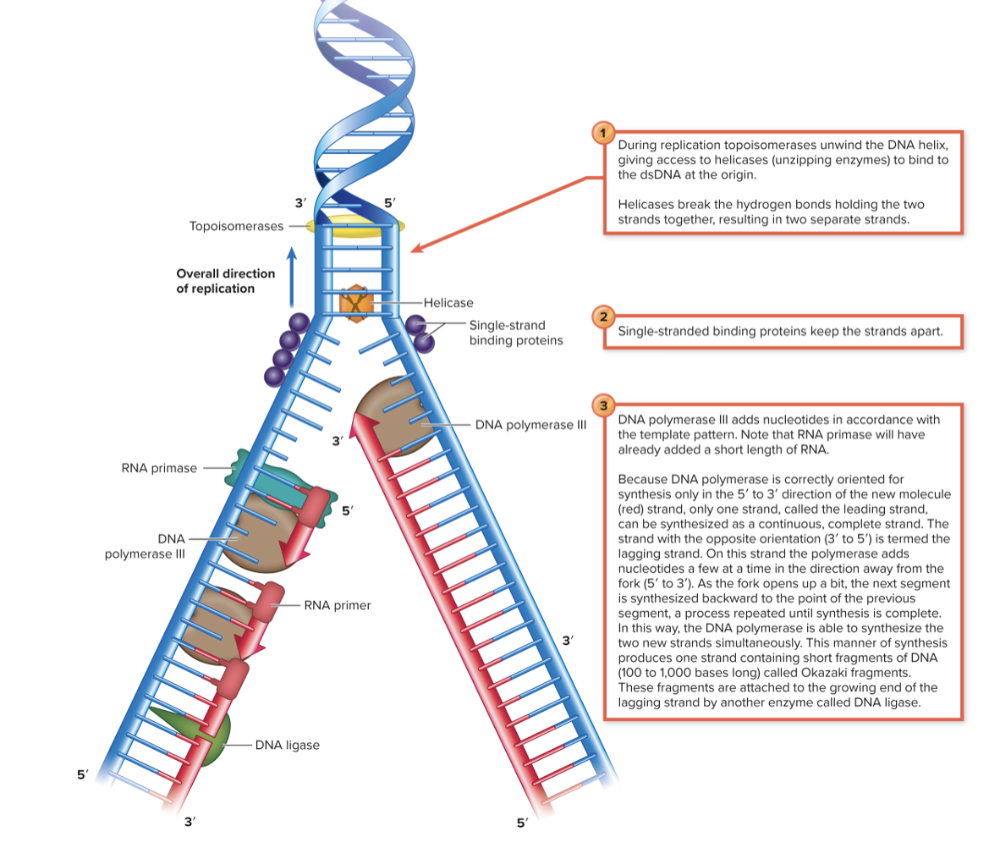 |
front 35 Explain elongation and termination of the daughter molecule in prokaryotes? | back 35 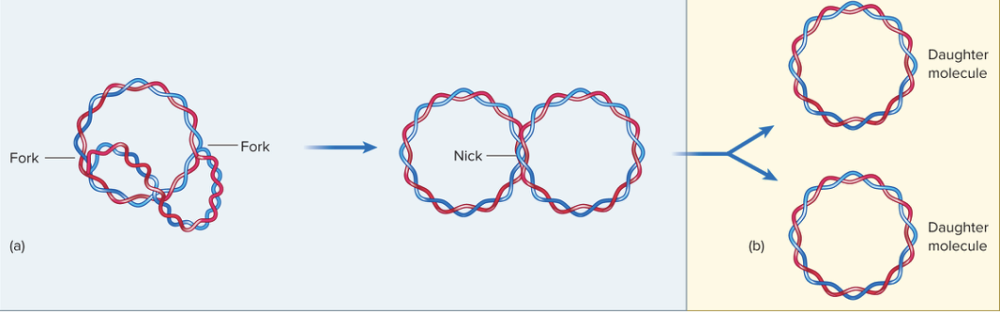 As DNA replication continues, the newly made DNA strands loop away from the original template because the replication forks move around the circular DNA molecule. In prokaryotes (like bacteria), the DNA is circular, so the two replication forks eventually meet on the opposite side of the circle—this is what it means when they “come full circle.” Once replication is almost finished, DNA ligase connects all the Okazaki fragments on the lagging strands, making each daughter DNA strand continuous and complete. Finally, Topoisomerase IV makes a temporary double-stranded break to separate the two new circular DNA molecules that are still linked together. After cutting, it reseals the DNA so that each daughter cell gets one complete, separate DNA molecule. |
front 36 Replication of eukaryotic DNA is similar to replication of _________ and ________ DNA. In what ways are they similar? | back 36 Replication of eukaryotic DNA is similar to replication of bacterial and archaeal DNA
|
front 37 Due to the unidirectional action of ___________, the 3′ end of linear DNA molecules, called _________, cannot be copied entirely | back 37 DNA polymerase telomeres |
front 38 Explain how the classical view of the “central dogma” of biology has been changed by recent science. | back 38 The classical view of the central dogma of biology states that information flows in one direction: DNA → RNA → protein. This means DNA is transcribed into RNA, and RNA is translated into proteins, which then carry out most cellular functions. However, recent science has shown that this view is oversimplified. Not all RNA molecules are used to make proteins. In fact, a large portion of our genome produces noncoding RNAs that have important roles on their own. These include rRNA and tRNA (which help in translation), as well as miRNA, siRNA, and lncRNA, which help regulate gene expression, control which genes are turned on or off, and even modify other RNAs. So today, we understand that the flow of genetic information is more complex: DNA can make RNA that doesn’t code for protein but still performs vital regulatory and structural functions. The modern view of the central dogma includes these noncoding RNAs as key players in controlling how genes are expressed and how cells function. |
front 39 What is the "central dogma" of genetics? | back 39 Transcription is when DNA is used to synthesize RNA Translation is when RNA is used for protein synthesis. However, the "central dogma" is incomplete because while a wide variety of RNAs are used to regular gene function. Many genetic malfunctions that cause human disease are found in regulatory RNA, not in genes for proteins. The DNA that codes for these crucial RNA molecules was once called "junk" DNA. |
front 40 ______________ regulate transcription and translation. | back 40 Micro RNA, interfering RNA, antisense RNA, and riboswitches |
front 41 A protein's _______ structure determines its characteristic shape and function | back 41 primary structure
|
front 42 _____________: the study of an organism’s complete set of expressed proteins | back 42
|
front 43 What is the simplified view of the DNA-Protein relationship | back 43 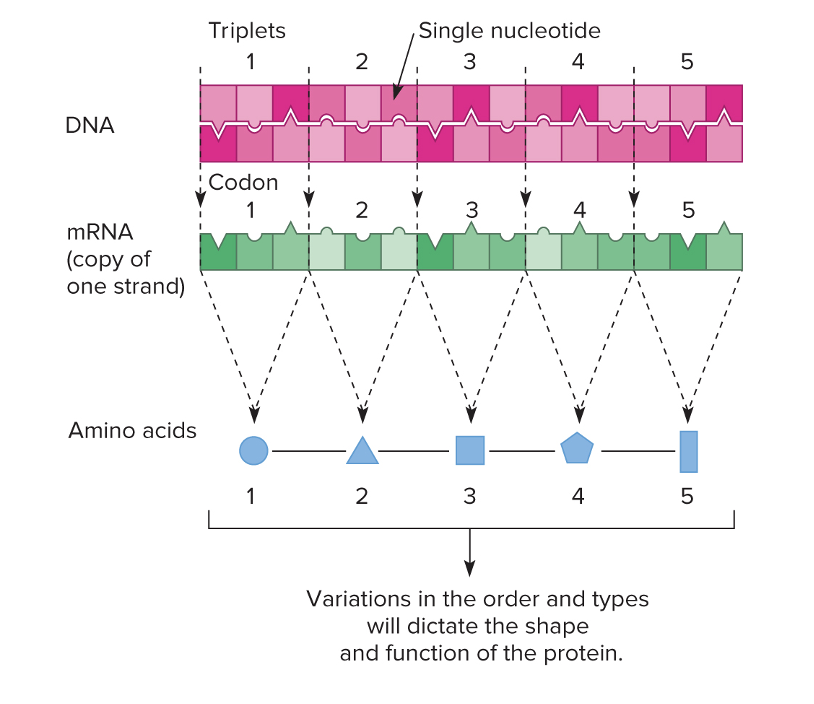 DNA codes into mRNA, and then three nucleotides make a codon. The tRNA carries the anti-codon which has the matching amino acid to the codon.
DNA coding strand: 5′ – ATG GCA TTT – 3′ |
front 44 What are the different participants in transcription and translation? | back 44 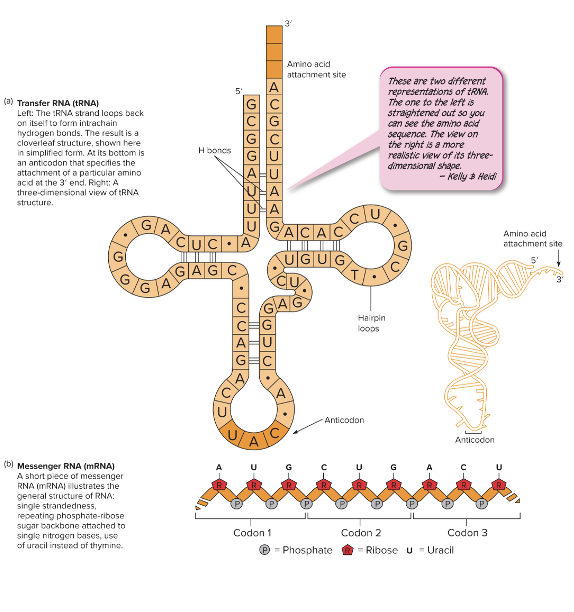
|
front 45 Which is the only type of RNA (ribonucleic acid) that can be translated? | back 45 mRNA This is a transcript of a structural gene or genes in the DNA. Synthesized in a process similar to the leading strand during DNA replication. |
front 46 Match the description to the type of RNA Sequence of amino acids in protein. Transports the DNA master code to the ribosome | back 46 mRNA |
front 47 Match the description to the type of RNA: A cloverleaf tRNA to carry amino acids. Brings amino acids to ribosome during translation | back 47 tRNA
|
front 48 Match the description to the type of RNA: Several large structural rRNA molecules. Forms the major part of a ribosome and participates in protein synthesis | back 48 rRNA It is a long RNA molecule and proteins form complex 3D shapes to create two subunits of a ribosome. The two subunits engage in the final translation of the genetic |
front 49 Match the description to the type of RNA: Regulation of gene expression and coiling of chromatin | back 49 Micro (miRNA), antisense, riboswitch, and small interfering (siRNA) Regulatory RNA’s. |
front 50 Match the description to the type of RNA: An RNA that can begin DNA replication. Primes DNA | back 50 Primer |
front 51 Match the description to the type of RNA: RNA enzymes, parts of splicer enzymes. Remove introns from other RNAs in eukaryotes | back 51 Ribozymes and spliceosomes (snRNA) |
front 52 What are codons? | back 52 A series of triplet bases that hold the message of the transcribed mRNA |
front 53 What are anticodons? | back 53 Found at the bottom of the cloverleaf. Designates the specificity of the tRNA and complements the mRNA codon |
front 54 Describe the process of transcription and what is involved? | back 54 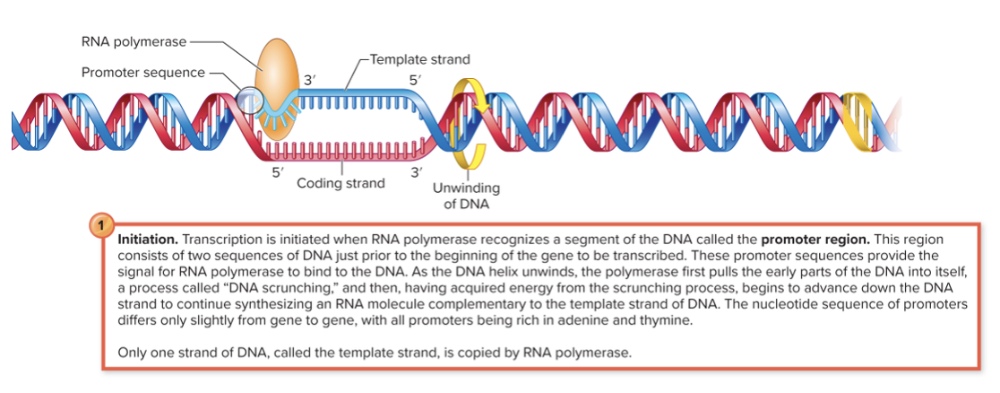 Initiation, Elongation, and Termination RNA polymerase binds to the promoter region of a gene. As it moves along the DNA, it unwinds a small section (about 10–20 base pairs at a time) by itself — no separate helicase enzyme is needed. It then uses one strand (the template strand) to synthesize RNA. The DNA re-anneals (zips back together) once RNA polymerase passes.
|
front 55 The genetic code is _____ for bacteria, archea, eukaryotes, and viruses. | back 55 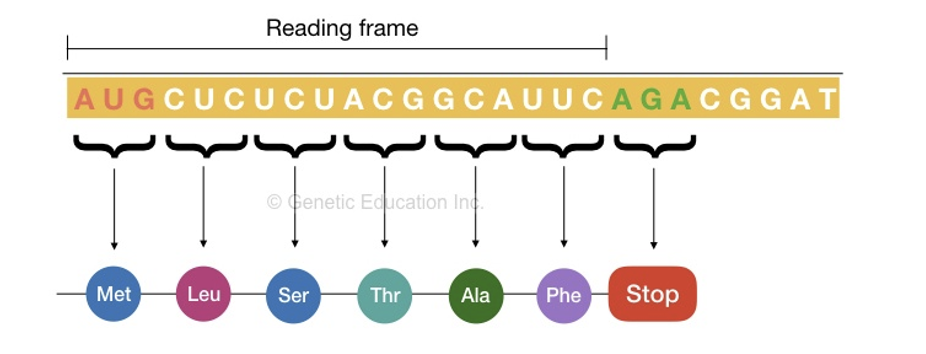 universal
|
front 56 What are the start codons? What are the stop codons? | back 56 AUG (Met); f'methionine in prokaryotes
Stop: UAA, UAG, UGA (these do not have an amino acid associated with them) |
front 57 Explain how the master genetic code is redundant and what is the wobble base? | back 57 Redundancy:
Wobble:
In the genetic code, several codons can code for the same amino acid—this is called redundancy or degeneracy of the genetic code. Because of this, if a mutation changes the third base in a codon (for example, from GAA to GAG), it often still codes for the same amino acid. This is known as the wobble effect. It helps protect cells, especially prokaryotes, from harmful mutations. Since the amino acid sequence of the protein doesn’t change, the structure and function of the protein stay the same, meaning the mutation doesn’t immediately harm or kill the cell. |
front 58 Interpreting the DNA code | back 58 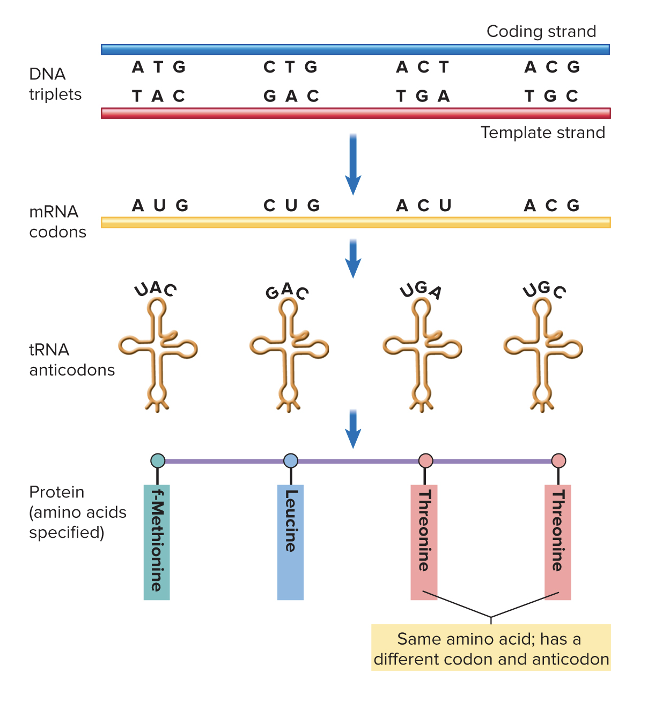 |
front 59 Describe the process of translation and what is needed. | back 59 mRNA, tRNA, amino acids, and ribosomes Three stages include initiation, elongation, and termination
Process Summary Translation is the process of making a protein from an mRNA sequence, and it occurs in three main stages: initiation, elongation, and termination. During initiation, the small ribosomal subunit binds to the mRNA strand. In prokaryotes, it binds near the Shine-Dalgarno sequence, while in eukaryotes, it attaches to the 5′ cap and scans for the start codon (AUG), which codes for methionine. A tRNA carrying methionine pairs with the start codon using its anticodon (UAC). Then, the large ribosomal subunit joins to form a complete ribosome with three sites: the A site (aminoacyl site, where new tRNAs enter), the P site (peptidyl site, where the growing peptide chain is held), and the E site (exit site, where tRNAs leave). During elongation, the ribosome moves along the mRNA in the 5′ to 3′ direction, reading one codon at a time. A new tRNA carrying an amino acid enters the A site, and the peptidyl transferase enzyme forms a peptide bond between the amino acid in the P site and the one in the A site. The ribosome then shifts, moving the tRNA with the growing chain into the P site, and the empty tRNA exits through the E site. This process repeats, adding amino acids one by one to the growing polypeptide chain. Finally, during termination, the ribosome reaches a stop codon (UAA, UAG, or UGA). Since there is no tRNA for these codons, a release factor binds to the A site, causing the ribosome to release the newly made protein. The ribosome then separates into its large and small subunits, and the mRNA and tRNA are released. The result is a completed polypeptide chain that will fold into its proper shape to become a functional protein. |
front 60 What are post-translational modifications? | back 60
|
front 61 Explain COVID-19 mRNA vaccines and how it uses translation. | back 61 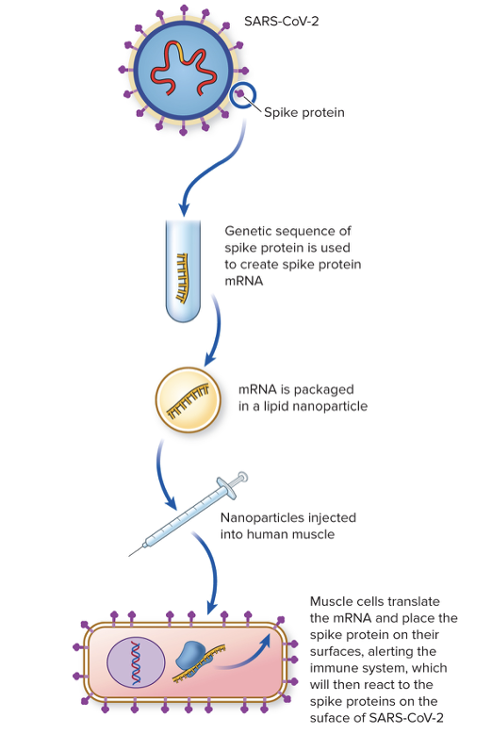 The COVID-19 vaccine uses viral mRNA that codes for a surface protein of the virus. Once injected into human areas, muscle cells use their translation machinery to create and place on their cell surface of the viral protein. This alerts the immune system, which will react to the spike proteins on surface of SARS-CoV2
|
front 62 Co-translational translation only occurs in _________ | back 62 Prokaryotes (bacteria and archea)
|
front 63 True or False: AUG codes for the same form of methionine in both prokaryotes and eukaryotes. | back 63 False; AUG codes for a different form of methionine in eukaryotes. |
front 64 Fill in the blanks: Eukaryotic mRNA’s only code for _______ protein; bacterial mRNA’s often contain information from _________ in a series, | back 64 one several genes Monocistronic = one → Eukaryotes → one gene per mRNA (one gene per protein) Polycistronic = many → Prokaryotes → many genes per mRNA |
front 65 In eukaryotic transcription and translation, what are introns and exons? | back 65 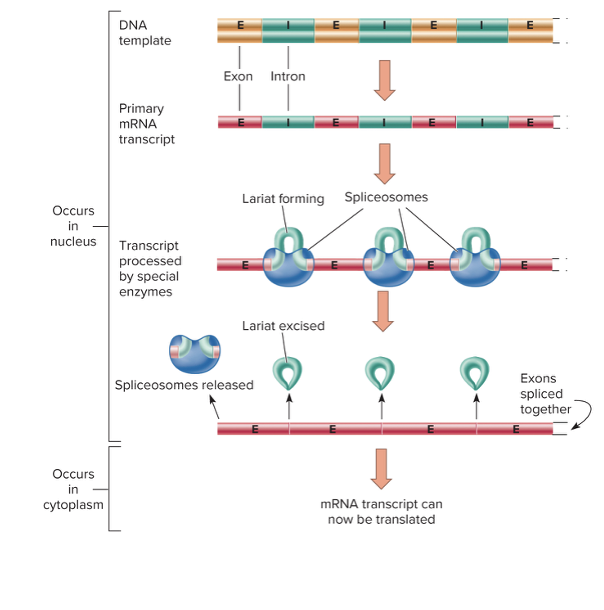 Most eukaryotic genes do not exist in an uninterrupted series of codons coding for a protein.
|
front 66 What is used to recognize the exon-intron junctions and enzymatically cut through them? | back 66 A spliceosome is used in eukaryotic translation and transcription. It loops introns into lariat-shaped pieces, excises them, and joins the exons end to end
|
front 67 Define the term operon and explain one advantage it provides to a bacterial cell. | back 67
________________________________ An operon is a group of genes that are controlled together and transcribed as a single mRNA molecule under the control of one promoter. These genes usually code for proteins that work in the same pathway or process. The main advantage of an operon is that it allows a bacterial cell to coordinate the expression of related genes all at once. This means the cell can efficiently turn on or off an entire set of genes in response to environmental changes — saving energy and resources. For example, in the lac operon, bacteria only produce lactose-digesting enzymes when lactose is present, which helps the cell avoid wasting energy making unnecessary proteins. |
front 68 What are inducible operons? | back 68 These operons contain genes that make proteins that help metabolize nutrients
|
front 69 What are repressible operons? | back 69 These operons contain genes that code for enzymes that synthesize amino acids. They are normally on, but can be turned OFF (repressed) when there is enough product synthesized by the enzyme and the nutrient is NO LONGER needed |
front 70 In repressible operons, excess nutrient serves as a _______ to block the action of the operon. | back 70 co-repressor
|
front 71 What is the Lac Operon? | back 71 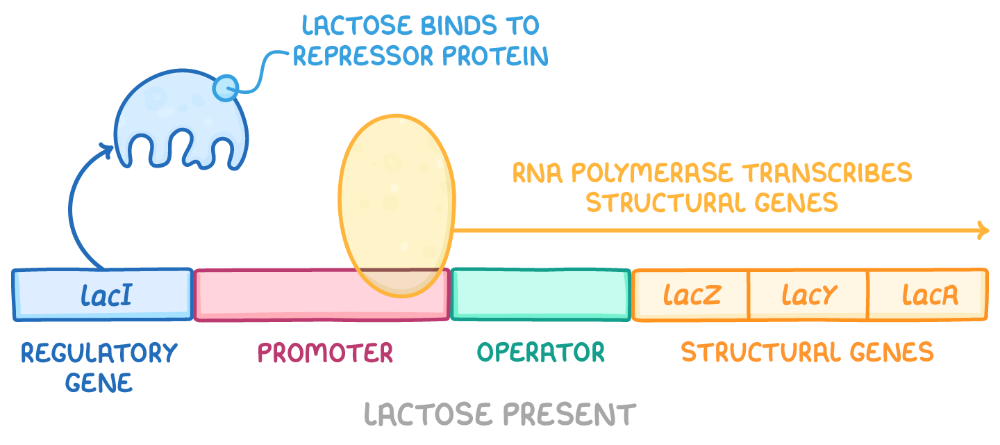 This operon is inducible, meaning it is off without lactose, but when lactose is present in the environment, it can be turned ON
|
front 72 The repressor protein is __________. What does this mean? | back 72 allosteric This means that it has two binding sites: one for the operator sequence on the DNA and another for the lactose. Allosteric binding is when a molecule binds to a protein at a site other than the active site, causing a change in the protein's shape that affects its function, either by increasing or decreasing its activity.
|
front 73 In the lac operon, the __________ causes a conformational change which allows transcription of the structural genes. | back 73 binding of allolactose to the repressor |
front 74 What is the Trp Operon? | back 74 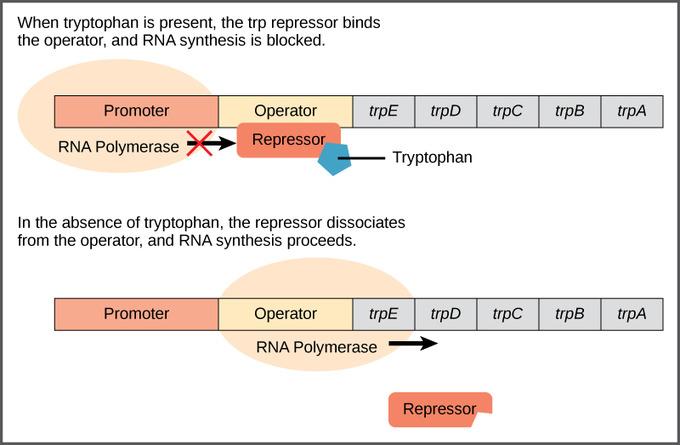 The trp operon is a group of genes in E. coli (and other bacteria) that control the production of the amino acid tryptophan. It contains five structural genes (trpE, trpD, trpC, trpB, and trpA), which code for the enzymes needed to synthesize tryptophan. These genes are all controlled by a single promoter and an operator region — together, this setup allows the cell to regulate tryptophan production efficiently. Here’s how it works:
In short, the trp operon is a repressible operon, meaning it’s usually on but can be turned off when the end product (tryptophan) is abundant. This allows bacteria to conserve energy by halting production of unnecessary enzymes when tryptophan is already available. |
front 75 What are two methods of eukaryotic gene regulation? | back 75
|
front 76 What are the four antibiotics that affect transcription and translation? ("mycins") RA SE | back 76
|
front 77 What is recombination? | back 77 Recombination is an event in which one bacterium DONATES DNA to another bacterium. The end result is a strain DIFFERENT from both the donor and recipient strain. |
front 78 What are plasmids? | back 78 Extrachromosomal DNA adept at moving between cells (small, circular pieces) Non-essential to the survival of the cell (but carry useful traits)
|
front 79 What is a recombinant? | back 79 It is any organism that contains and expresses genes that originated in another organism. |
front 80 What is horizontal gene transfer in bacteria, and what are the three examples? | back 80 Any transfer of DNA that results in organisms acquiring NEW genes that did not come DIRECTLY from the parent organisms.
|
front 81 What is the ONE type of DIRECT horizontal gene transfer? | back 81 Conjugation, and direct meaning the donor and recipient are IN CONTACT during the exchange |
front 82 What are the two types of INDIRECT horizontal gene transfer? | back 82 Transformation and Transduction |
front 83 What is conjugation? | back 83 Factors Involved:
Examples of Products of Transferred Genes
What is it?
|
front 84 Explain the process of conjugation with the F factor. | back 84 Conjugation is a process in which one bacterium transfers genetic material to another through direct contact. It requires a special piece of DNA called the F factor (fertility factor). The F factor is a plasmid, which means it is a small, circular piece of DNA separate from the bacterial chromosome. (F factor is from plasmid NOT bacterial chromosome) Conjugation occurs between a donor cell (F⁺), which contains the F factor, and a recipient cell (F⁻), which does not. The F⁺ donor forms a thin bridge-like structure called a sex pilus that attaches to the F⁻ recipient. The pilus then pulls the two cells together, forming a direct connection. Once connected, the donor cell begins to copy its F factor plasmid. As replication occurs, one strand of the F factor DNA is transferred through the pilus into the recipient cell. Both cells then synthesize the complementary strand of the DNA, so each ends up with a complete F factor plasmid. After the transfer, the recipient becomes F⁺, meaning it can now act as a donor in future conjugation events. In summary, conjugation with the F factor allows bacteria to share genetic material, spreading traits like antibiotic resistance or new metabolic abilities — all without reproduction. |
front 85 What are resistance plasmids in conjugation? | back 85 Resistance plasmids or factors
|
front 86 What experiment discovered transformation? | back 86 
|
front 87 What is transformation? | back 87 Factors Involved
Examples of Products of Transferred Genes:
What is it?
|
front 88 What is transduction? | back 88 Factors Involved
Examples of Products of Transferred Genes
What is it?
|
front 89 Participating bacteria in a single transduction event must be the ______ species. The two versions are: | back 89 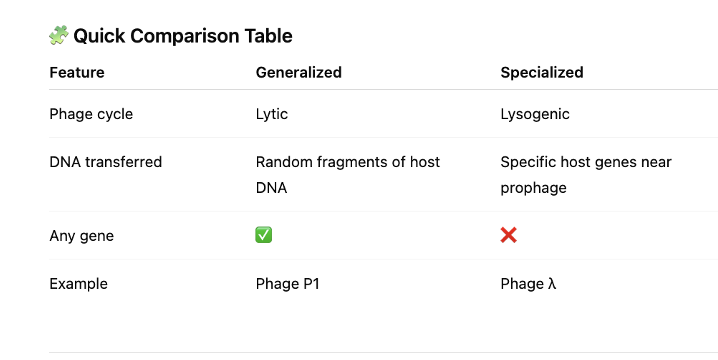 - same species Generalized transduction
Specialized transduction
|
front 90 What are transposons or transposable elements (TEs)? | back 90 "Jumping genes" shift from one part of the genome to another
Proposed by Barbara McClintock Widespread among cells and viruses
|
front 91 What are the different types of transposable elements? | back 91 Transposons are pieces of DNA that can move around inside a cell’s genome. Insertion elements:
Retrotransposon:
Other
|
front 92 What are the general effects of transposable elements? | back 92 Mix up genetic language Can be beneficial or adverse, depending on:
|
front 93 What are the effects of transposable elements on bacteria? | back 93
|
front 94 What are pathogenicity islands (PAI's)? | back 94 Pathogenicity islands (PAIs) are special sections of DNA found in some bacteria. These sections contain multiple genes that work together (coordinate) to give the bacteria a new ability/trait that makes it harmful (pathogenic). For example:
These DNA “islands” are often surrounded by transposon-like sequences, which means they may have been moved or transferred between bacteria in the past. In short, pathogenicity islands are clusters of genes that can turn harmless bacteria into disease-causing ones by adding new traits like toxin production or nutrient stealing. Islands flanked by sequences that look like genes for TE enzymes
|
front 95 What is a mutation? | back 95 Any change to the nucleotide sequence in the genome
|
front 96 What is the difference between the wild type vs mutant strain? | back 96 Wild type: (original)
Mutant strain:
|
front 97 What are the two different causes of mutations? | back 97
|
front 98 What are mutagens? | back 98 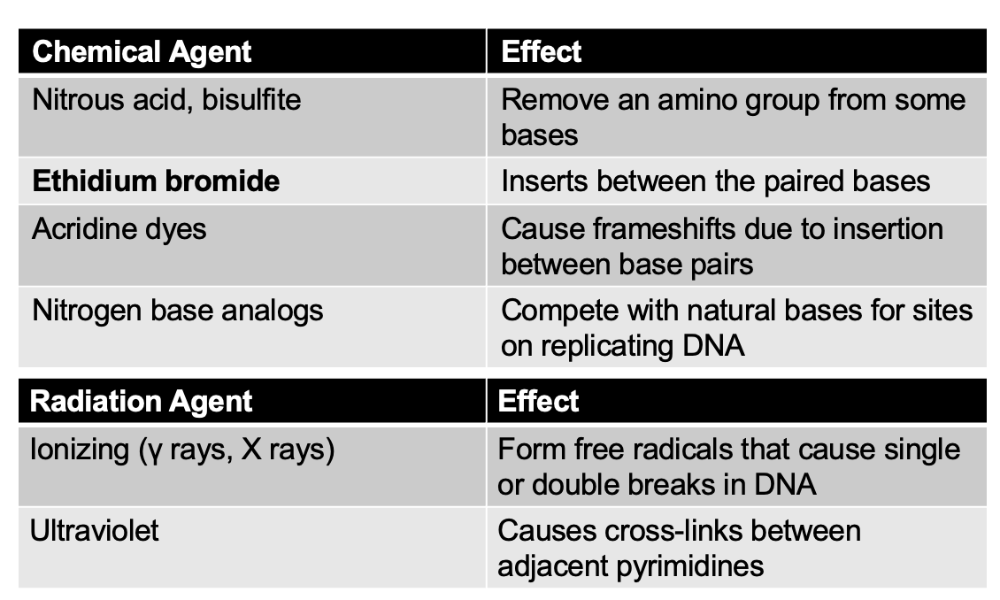 Physical and chemical agents that damage DNA leading to induced mutations |
front 99 Three overarching categories of mutations: Point, Lethal, and Neutral. What do they mean | back 99 Point mutations:
Lethal mutation:
Neutral mutation
|
front 100 What is a missense, nonsense, silent, and back mutation? | back 100 Missense mutation: (changing amino acid)
Nonsense mutation:
Silent mutation:
Back mutation:
|
front 101 What is a frameshift mutation? | back 101
|
front 102 With damage caused by mutations, what are two ways to fix the repair? | back 102 1. Photo-activation (non-ionizing)
2. Mismatch Repair
|
front 103 What is excision repair of mutations, and how is it different from mismatch repair? | back 103
|
front 104 What is the Ames Test? | back 104 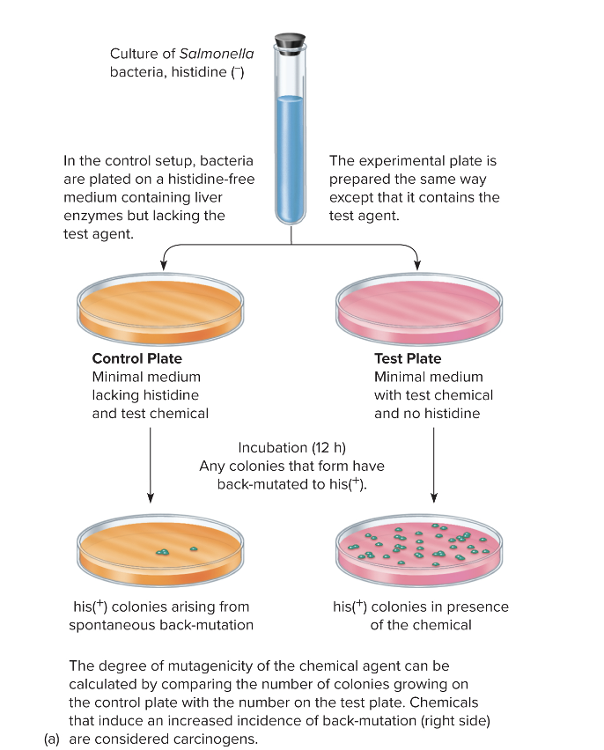
The Ames test is a lab test used to see if a chemical can cause mutations. Scientists care about this because mutagens often end up being carcinogens. The test uses bacteria (Salmonella) that have a specific mutation preventing them from making histidine, which is an amino acid they need to grow. Because they can’t make histidine, these bacteria cannot grow on a plate that doesn’t have histidine in it — unless a new mutation “fixes” the original mutation. This “fix” is called a reversion mutation. The goal of the Ames test is to see whether a chemical causes more of these reversion mutations than would normally happen by chance. Here’s how it works in a simple way:
So basically: more bacterial growth after chemical exposure = the chemical is likely a mutagen. |
front 105 What are some of the positive and negative effects of mutations? | back 105
|
front 106 Chapter 21 | back 106 Infectious Diseases Manifesting in the Cardiovascular and Lymphatic Systems |
front 107 What are the different functions and features of the cardiovascular system? | back 107
|
front 108 What are arteries? | back 108 Carry blood away from the heart Arterioles - smaller branches |
front 109 What are veins? | back 109 Carry blood toward the heart Venules: smaller veins |
front 110 What are capillaries? | back 110 Smallest blood vessel Connect venules and arterioles |
front 111 What are the components and functions of the lymphatic system? | back 111
|
front 112 What are the defenses of the cardiovascular system? | back 112
|
front 113 What are the defenses of the lymphatic system? | back 113
Lymph nodes filter lymph fluid and trap anything foreign—like pathogens, debris, or abnormal cells—so immune cells can destroy them. Inside lymph nodes, you have dense populations of lymphocytes (B cells and T cells), macrophages, and dendritic cells that identify antigens and mount immune responses. The flow of lymph itself also acts as a surveillance mechanism, constantly moving material from tissues to nodes where it can be inspected. Additionally, the lymphatic system contains the spleen, which filters blood and helps initiate immune responses, and the thymus, which produces T cells. The system also includes structural defenses like valves that help maintain one-way flow, preventing stagnation where pathogens could accumulate. So instead of microbiota, the lymphatic system is defended by an organized network of immune tissues and cells built specifically to detect and eliminate threats. |
front 114 Medical conditions involving blood have the suffix _____ | back 114 -emia. |
front 115 What is viremia? | back 115 Viruses that cause meningitis |
front 116 What is fungemia? | back 116 Fungi in the blood |
front 117 What is bacteremia? | back 117 Presence of bacteria in the blood |
front 118 What is septicemia? | back 118 Bacteria that are flourishing and growing (actively reproducing) in the bloodstream This can lead to decreased blood pressure and septic shock. |
front 119 True or False: The cardiovascular, lymphatic, and nervous system are closed, which means there is no normal biota. Microorganisms can be present transiently. | back 119 True NO normal microbiota It means that microbes may enter or appear in a system for a short period of time, but they do not stay, do not colonize, and do not become part of the normal microbiota. They’re just “passing through.” Recent data from microbiome studies:
|
front 120 What is COVID-19 caused by? | back 120
|
front 121 What are the different variants of the SARS-CoV-2? | back 121
|
front 122 COVID-19 is caused by, and what are its other characteristics? | back 122 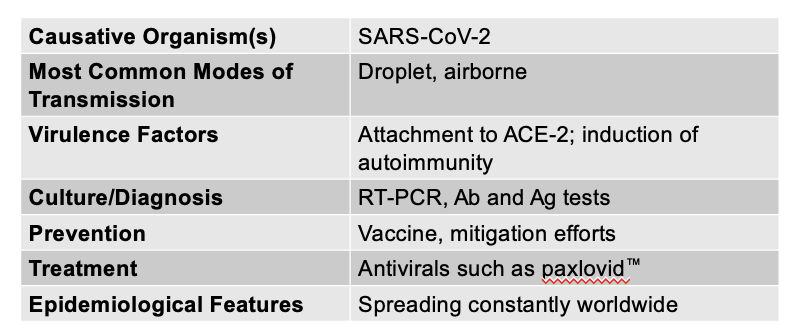 Prominent respiratory symptoms
|
front 123 Endocarditis is caused by, and what are its other characteristics? (Main information - there are two subtypes) | back 123
|
front 124 What is parenteral mode of transmission? | back 124 Parenteral transmission means a microorganism enters the body through a route other than the digestive tract, usually by punctures, injections, bites, cuts, or any break in the skin or mucosa. This includes things like needle sticks, IV drug use, contaminated medical equipment, animal bites, or traumatic injuries. The key idea is that the pathogen bypasses the normal protective barriers (skin and mucous membranes) and is delivered straight into deeper tissues or the bloodstream, which makes infection easier. |
front 125 Acute endocarditis is caused by, and what are the other characteristics? | back 125 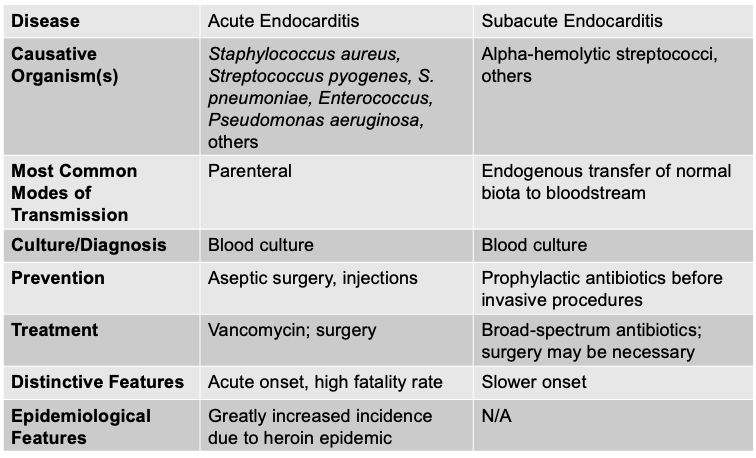 - Fever, anemia, abnormal heartbeat, symptoms of heart attack, SOB, chills - Abdominal/side pain, JANEWAY lesions (painless, red or purple spots on the palms and soles), Osler's nodes (small, tender, raised, red or purple nodules that typically appear on the pulp of the fingers and toes) - Acute endocarditis is a fast-moving, aggressive infection of the heart valves, usually caused by highly virulent organisms like Staphylococcus aureus. It can destroy valves within days, cause high fevers, sepsis, emboli, and is immediately life-threatening.
|
front 126 Subacute endocarditis is caused by, and what are the other characteristics? | back 126  - Similar to symptoms of acute endocarditis - Develop more slowly and are less pronounced - Enlarged spleen, clubbed fingers, and toes (Endocarditis causes an enlarged spleen (splenomegaly) because the spleen is constantly filtering infected blood and trapping immune complexes, bacteria, and debris. The ongoing immune activation makes the spleen work overtime, causing it to swell. The clubbed fingers and toes come from long-standing inflammation that leads to increased blood flow and growth of soft tissue at the fingertips) - Subacute endocarditis, on the other hand, is slower and less aggressive, usually caused by low-virulence organisms like viridans streptococci. It tends to develop over weeks to months, often on previously damaged valves, and symptoms are milder and more gradual.
|
front 127 Does both gram- and gram + produce endotoxins? (Extra) | back 127 Endotoxin refers specifically to lipid A, the toxic portion of lipopolysaccharide (LPS), which is a major component of the outer membrane of Gram-negative bacteria. Gram-positive bacteria do not have an outer membrane at all, so they cannot produce endotoxin. Instead, Gram-positive bacteria may release other inflammatory molecules—like teichoic acids or superantigens—but these are not classified as endotoxins. |
front 128 Sepsis is caused by, and what are the other characteristics? | back 128 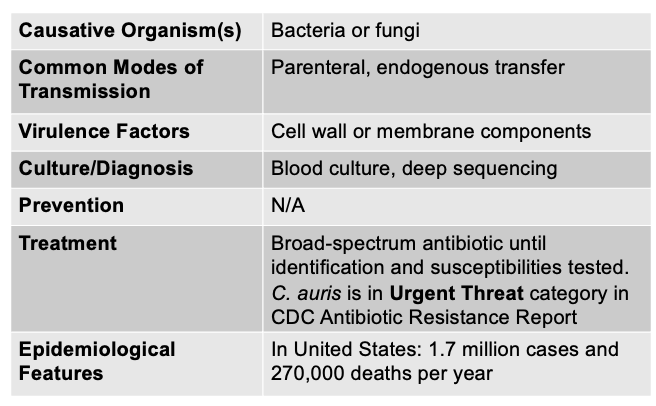 - also called septicemia - occurs when organisms are actively multiplying in the blood (can be caused by many different bacteria and some fungi) - Signs/Symptoms: fever, AMS, shaking, chills, GI symptoms, increased breathing rate, respiratory alkalosis, low BP resulting in loss of fluid from vasculature
|
front 129 What is endotoxic shock? | back 129
Sepsis is a broad, body-wide inflammatory response to an infection—bacterial, viral, or fungal—that leads to organ dysfunction. It happens when the immune system overreacts to an infection and releases massive amounts of cytokines, causing fever, low blood pressure, clotting abnormalities, and organ damage. Endotoxic shock, on the other hand, is a subset of septic shock that occurs specifically during infections with Gram-negative bacteria that release endotoxin (lipid A from LPS). This endotoxin triggers an extreme immune reaction that causes severe vasodilation, a dangerous drop in blood pressure, DIC, and multi-organ failure. In other words, all endotoxic shock is sepsis, but not all sepsis is endotoxic shock—because sepsis can be caused by pathogens that don’t have endotoxin, like Gram-positive bacteria, fungi, or viruses. |
front 130 What are the three manifestations of the plague? (PBS) | back 130 1. Pneumonic plague
2. Bubonic plague
3. Septicemic plague
|
front 131 For the bubonic plague, the number of bacteria required to initiate infection in bubonic (or septicemic) cases is only ______ to ____ cells. | back 131  3 to 50 cells |
front 132 Plague Disease is caused by, and what are the other characteristics | back 132 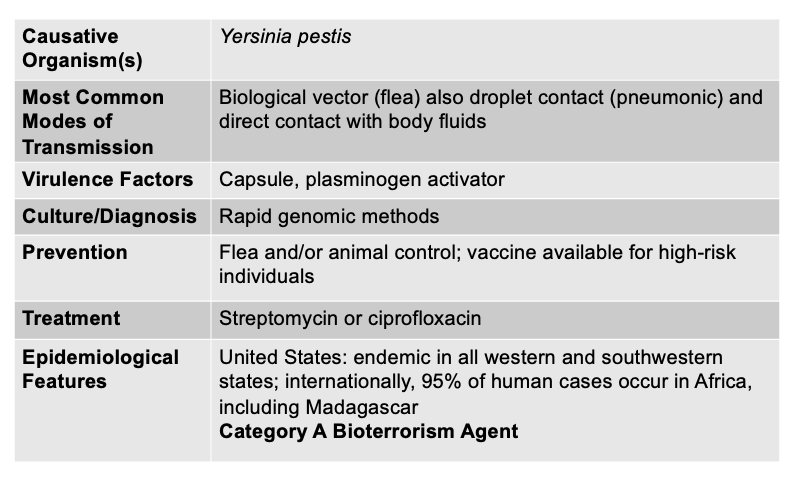 All three classical forms of plague (bubonic, septicemic, and pneumonic) are caused by the same bacterium: Yersinia pestis.
|
front 133 What are some characteristics about Lyme Disease? | back 133
|
front 134 What is the early symptom of lyme disease? | back 134 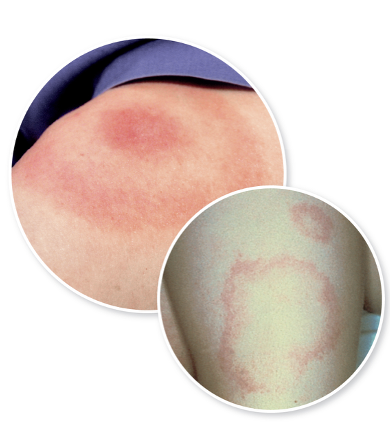
|
front 135 What does lyme disease develop into after several weeks and months? | back 135
|
front 136 Lyme Disease is caused by what, and what are the unique characteristics | back 136 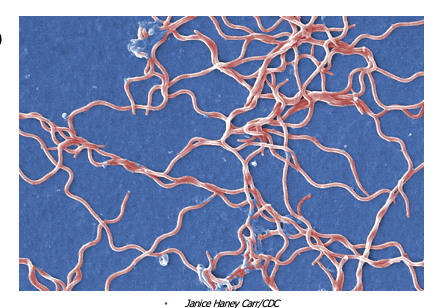 - Caused by a large spirochete with 3-10 irregularly spaced coils - Evades the immune system by changing antigens (antigenic variation, where it repeatedly changes the surface proteins displayed on its outer membrane. Because antibodies are specific to a particular protein shape, these shifts prevent the immune system from recognizing and eliminating the bacteria effectively. This constant “costume change” allows the organism to persist in the body for months to years, contributing to the chronic, relapsing nature of Lyme disease symptoms.) - Has multiple proteins for attachment to host cells (The bacterium produces a variety of outer surface proteins (Osps) that bind to different host tissues. Some help it attach to tick midguts, some help it enter the bloodstream, and others bind to human cells like fibroblasts, endothelial cells, and components of the extracellular matrix such as collagen. This wide range of adhesion proteins allows Borrelia to spread through the skin, joints, nervous system, and heart) - Possible that immune response contributes to the pathology of the disease
|
front 137 Infectious mononucleosis is caused by what, and what are the unique characteristics | back 137 - Epstein-Barr virus
Causative organisms: Epstein-Barr virus Mode of Transmission: Direct, Indirect, parenteral Virulence: latency, ability to incorporate into host DNA Culture/Diagnosis: differential blood count, MONOspot test for heterophile antibody, specific ELISA Prevention: N/A Treatment: Supportive Distinctive Features: Lifelong persistence Epidemiological Features: 500 cases per 100,000 per year there’s no treatment for EBV because antiviral medications don’t work well against it, and the illness is mostly caused by your immune system reacting to the virus—not the virus actively replicating at high levels. Once EBV infects B-cells, it quickly enters a latent state, meaning it hides inside cells with almost no active replication. Antivirals can only target viruses that are actively making new copies, so they have virtually no effect on a virus that is lying dormant. Because the body’s immune system eventually controls the acute infection on its own, treatment focuses on rest, hydration, and supportive care rather than killing the virus directly. Even though EBV establishes lifelong persistence, we don’t keep getting sick repeatedly because the virus stays in a controlled, latent phase within a small population of memory B-cells. In this state, the immune system—especially cytotoxic T-cells—constantly monitors and suppresses it. EBV only reactivates occasionally, and when it does, the immune system shuts it down before symptoms appear. That’s why you don’t keep getting “mono” again and again, and most reactivations are completely silent unless a person becomes severely immunocompromised. The immune system’s long-term surveillance keeps EBV from causing repeated illness despite its permanent presence. |
front 138 Anthrax is caused by what, and what are the unique characteristics | back 138 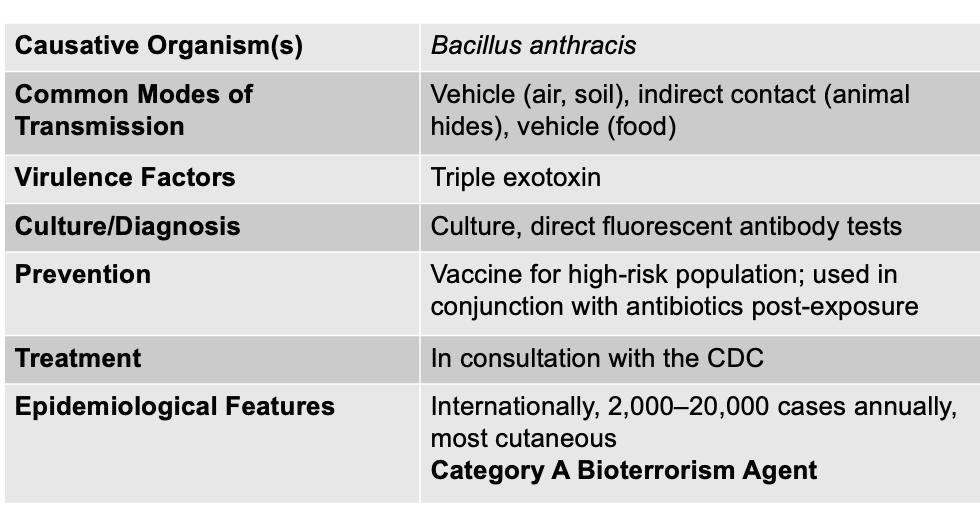 - Bacillus anthracis (+), endospore forming rod - Aerobic and catalase positive - Forms a tripartite toxin (PEL) Causative agent: Bacillus anthracis Mode of Transmission: Vehicle (air, soil), indirect (animal hides), and vehicle (food) Virulence Factor: tripartite toxin (triple exotoxin) Culture/Diagnosis: Culture; direct fluorescent antibody tests Prevention: Vaccine for high-risk population; used in conjunction with antibiotics post-exposure Treatment: in consultation with CDC Epidemiological Features: Internationally, 2,000–20,000 cases annually, most cutaneous; Category A Bioterrorism Agent |
front 139 Bacillus anthracis, an aerobic gram + endospore forming rod bacterium forms a tripartite toxin, which includes? | back 139  (PEL) Protective antigen Edema Factor Lethal Factor Bacillus anthracis forms a tripartite toxin, meaning it uses three separate proteins—protective antigen, edema factor, and lethal factor—that work together to damage host tissues. Protective antigen binds to host cells and creates a channel that allows the other two toxins inside. Edema factor increases cyclic AMP levels, causing massive fluid leakage and swelling. Lethal factor disrupts signaling pathways in immune cells, leading to cell death and systemic toxicity. None of the components are very harmful alone, but together they create the powerful anthrax toxin that causes severe edema, immune system collapse, and tissue damage. |
front 140 What are hemorrhagic fevers? | back 140 These are agents that infect the blood and lymphatics
|
front 141 Yellow Fever is caused by what, and what are the unique characteristics? | back 141 
Causative Agent: Yellow fever virus Mode of Transmission: Biological vector (Aedes mosquito) Virulence Factor: Disruption of clotting factors
Culture/Diagnosis: ELISA/PCR Prevention: Live attenutated vaccine available Treatment: Supportive Distinctive Features: accompanied by jaundice Epidemiological Features: only sporadic cases in travelers; international, 200,000 cases annually, 30,000 deaths; 90% of cases in Africa |
front 142 What are non-hemorrhagic fevers? | back 142
|
front 143 Cat-Scratch disease is caused by what, and what are the unique characteristics? (NH) | back 143  - Bartonella henselae
Bartonella henselae Mode of Transmission: Parenteral (cat scratch or bite) Virulence Factor: Endotoxin ( endotoxin, which is a type of lipopolysaccharide (LPS) found on the outer membrane of all Gram-negative bacteria. This endotoxin triggers a strong inflammatory response when the bacteria enter the skin through a scratch or bite. The inflammation causes the characteristic swollen, tender lymph nodes (lymphadenitis). Bartonella also invades endothelial cells and can survive inside immune cells, which helps it avoid clearance. But the endotoxin is what drives much of the fever, malaise, and lymph node swelling because it activates the immune system and promotes localized inflammation.) Culture/Diagnosis: Biopsy of lymph nodes plus Gram staining; ELISA (performed by CDC) Prevention: Clean wound sites Treatment: Azithromycin or rifampin Distinctive Features: History of cat bite or scratch; fever not always present Epidemiological: United States: estimated incidence is 9.3 cases per 100,000; internationally, seroprevalence from 0.6% to 37% depending on cat population |
front 144 Spotted Fever Rickettsiosis is caused by what, and what are the unique characteristics? (NH) | back 144  - Rocky Mountain Spotted Fever (RMSF) is most well-known - Mainly occurs in the southeast and eastern seaboard regions of the U.S, Canada, Central/South America
- Signs/symptoms: chills, fevers, headache, muscular pain - Distinctive RASH occurs 2-4 days after prodromal symptoms - If untreated, lesions merge and become necrotic - Cardiovascular disruption: hypotension, thrombosis, hemorrhage 20% mortality if untreated, 5-10% mortality if treated Causative agent: Rickettsia species Mode of Transmission: Biological vector (tick) Virulence Factors: Induces apoptosis in cells lining blood vessels (Because apoptosis does not tear or rupture the blood vessel wall, the vessels remain structurally intact. This means blood does not leak out into tissues, so you don’t get hemorrhage, bruising, or bleeding.) - causing these blood vessel–lining cells to undergo programmed cell death, the pathogen weakens the barrier enough to impair normal function—reducing blood flow, altering nutrient delivery, and helping the pathogen access deeper tissues—while avoiding the loud immune alarm that necrosis would cause. Culture/Diagnosis: Fluorescent antibody, PCR Prevention: Avoid ticks Treatment: Doxycycline Distinctive Features: Rocky Mountain spotted fever is most severe of the rickettsioses Epidemiological: Only in Americas; 10-fold increase since 2000 |
front 145 What are some basic characteristics about HIV? | back 145 Human Immunodeficiency Virus
|
front 146 HIV causes _________? | back 146
|
front 147 What are the complex of symptoms caused by HIV? | back 147 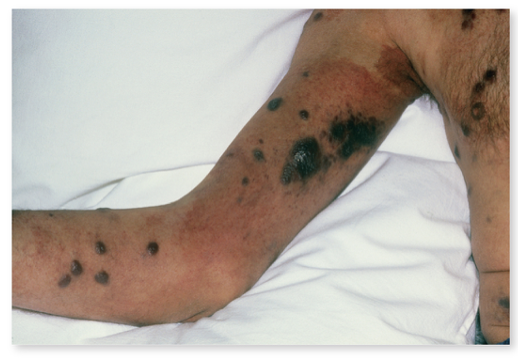
|
front 148 Signs and symptoms of HIV/AIDS are tied to the level of _____ in the blood and the level of ____ in the blood | back 148 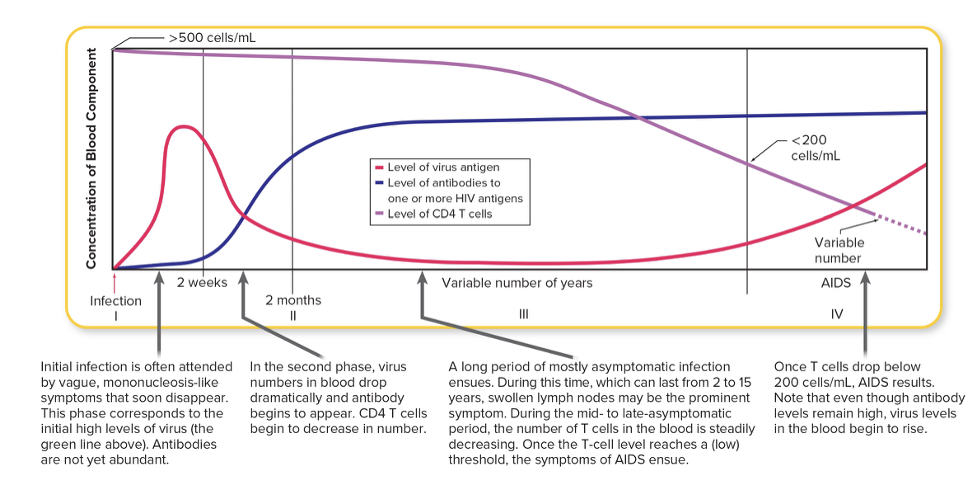
|
front 149 What are the different stages of HIV/AIDs disease progression? | back 149 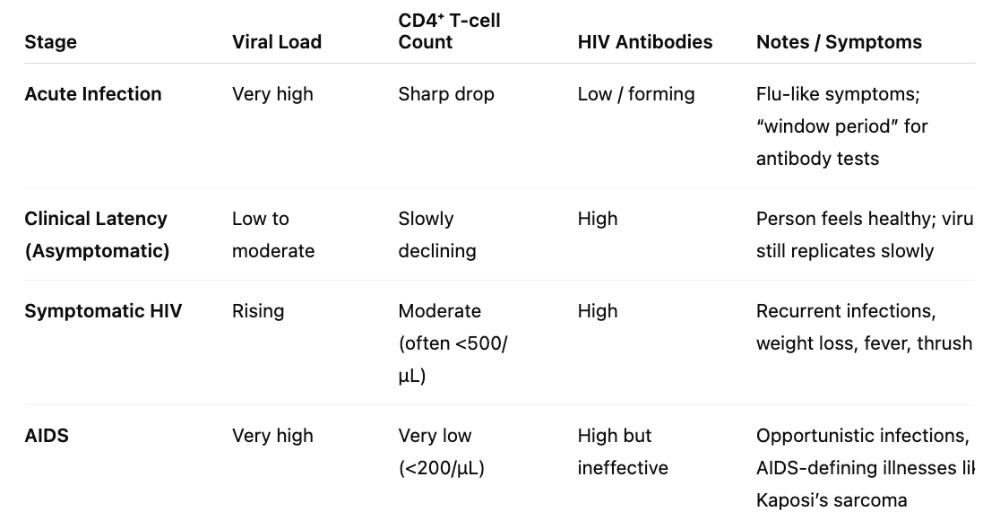 |
front 150 What are additional signs/symptoms for HIV Infection/AIDS? | back 150
|
front 151 What is the causative agent of HIV Infection/AIDs? | back 151
Two major types of HIV:
|
front 152 What is the general multiplication cycle of HIV? | back 152 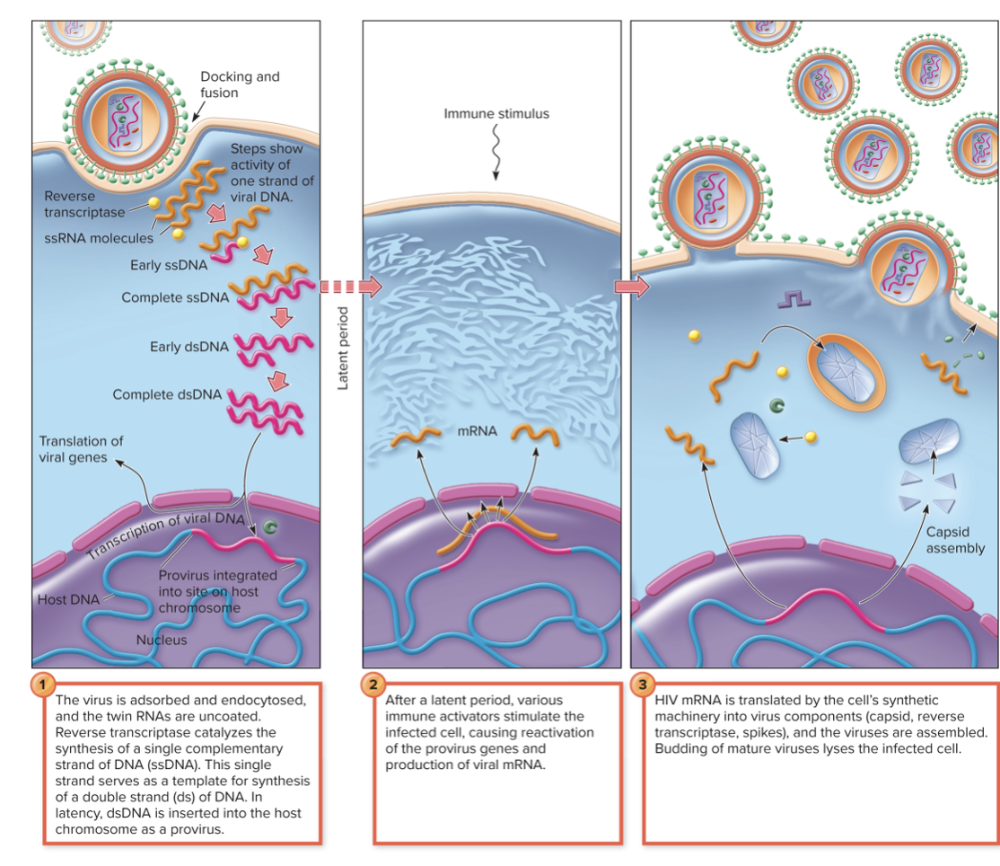
So the immune stimulus acts like an alarm that wakes up latent HIV, allowing it to replicate and spread again. |
front 153 For HIV transmission, (T/F), urine, tears, sweat, and saliva are considered sources of infection. | back 153 FALSE |
front 154 For HIV transmission, what is considered forms or sources of transmission? | back 154 Any form of intimate contact involving transfer of blood can be a potential source of infection
|
front 155 What is the culture and diagnosis for HIV? | back 155 A person is diagnosed as having HIV infection if they tested POSITIVE for exposure to HIV (diagnosis is NOT the same as having AIDS)
|
front 156 What are the two criteria that diagnosis of AIDS is based on? | back 156 Diagnosis with Stage 3 HIV infection requires both:
AIDS is the most advanced stage of HIV infection. It’s diagnosed when CD4⁺ T-cell counts drop below 200/µL or when the person develops AIDS-defining illnesses like Kaposi’s sarcoma, Pneumocystis pneumonia, or other opportunistic infections. So Stage 3 HIV = symptomatic but not always AIDS, whereas AIDS = end-stage HIV with severe immune compromise. AIDS, on the other hand, is not a virus—it’s a syndrome, meaning a collection of conditions caused by advanced HIV infection. To determine if someone has progressed to AIDS, doctors look at CD4⁺ T-cell counts and check for AIDS-defining illnesses. |
front 157 What are some prevention strategies for HIV Infection? | back 157
|
front 158 What is the treatment plan for HIV infection and AIDS? What is the three drug cocktail? | back 158
|
front 159 What are the three main mechanisms of action for Anti-HIV drugs? | back 159 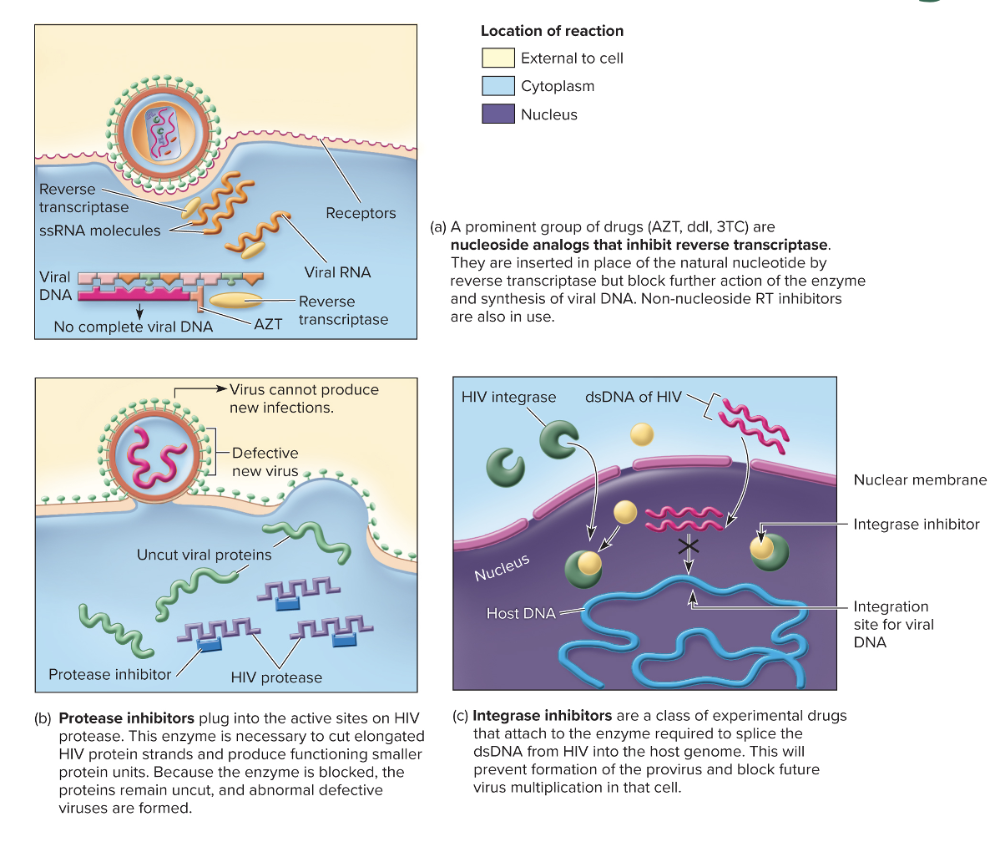
|
front 160 FINALLY ... HIV Infection and AIDS is caused by what, and what are the unique characteristics? | back 160 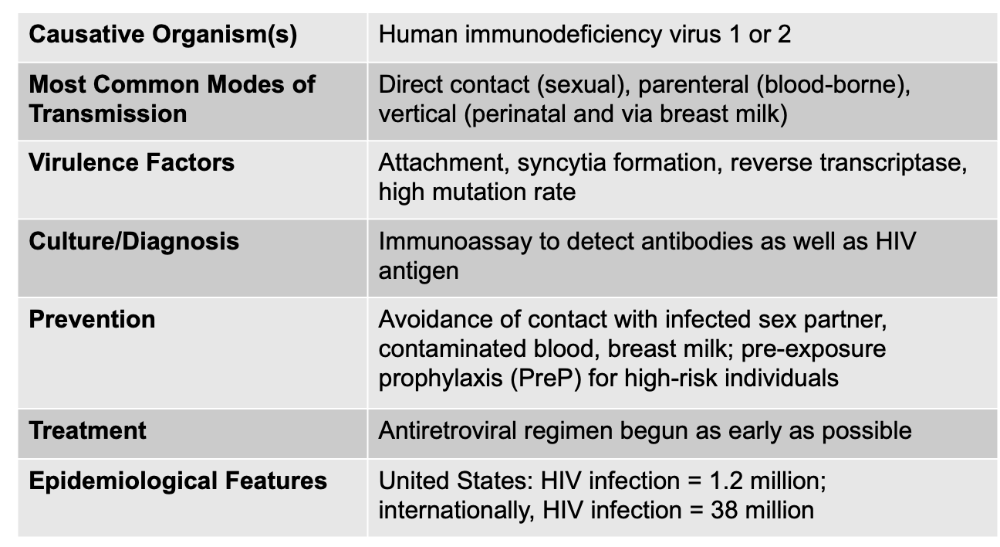 Virulence Factors: Attachment, syncytia formation, reverse transcriptase, high mutation rate
* PreP (pre-exposure prophylaxis) for high-risk individuals |
front 161 Bacillus anthracis is .... | back 161 Gram Positive Endospore Forming Bacteria Aerobic Rod-Shaped Anthrax |
front 162 Staphylococcus aureus is .. | back 162 Gram Positive Acute Endocarditis |
front 163 Streptococcus pneumoniae | back 163 Gram Positive diplococci joined end to end Acute Endocarditis Virulence: Capsule OR hemolysin
|
front 164 Yersinia pestis is .. | back 164 Gram Negative Plague (Pneumonic, Bubonic, Septicemic) |
front 165 Borrelia burgdorferi is | back 165 Gram Negative Lyme Disease |
front 166 Bartonella henselae is | back 166 Gram Negative Cat-Scratch Disease |
front 167 Rickettsia species is | back 167 Gram Negative Spotted fever rickettsiosis |
front 168 Epstein-Barr Virus is ... | back 168 DNA virus Infectious mononucleosis |
front 169 SARS-CoV2 is ... | back 169 RNA virus COVID-19 |
front 170 Yellow Fever virus is... | back 170 RNA virus Yellow Fever |
front 171 Human immunodeficiency virus 1 and 2 is a ... | back 171 Retrovirus HIV infection and AIDS |