BMD 320: Exam 3 Learning Objectives
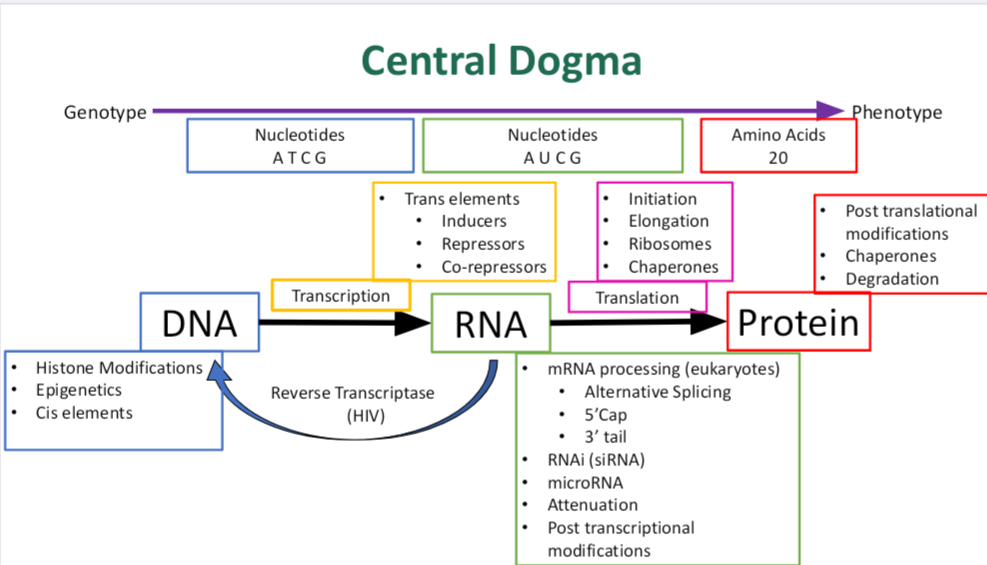
•What is the central dogma?
describes the flow of genetic information in a cell:
DNA → RNA → Protein
Breakdown: DNA is transcribed into RNA. RNA is translated into protein.
This explains how the instructions in DNA are used to build proteins, which carry out most cellular functions.
Bonus: There are exceptions, like reverse transcription in retroviruses (e.g., HIV), where RNA → DNA.
•What are the main steps?
The main steps of the central dogma are:
1. Replication: DNA makes a copy of itself.
Enzyme: DNA polymerase
2. Transcription: DNA is used to make messenger RNA (mRNA).
Enzyme: RNA polymerase
3. RNA Processing (in eukaryotes only): mRNA is spliced, capped, and gets a poly-A tail.
4. Translation: mRNA is read by ribosomes to build a protein (a chain of amino acids).
In short: Replication → Transcription → (RNA Processing) → Translation
•What molecules play an important role in the central dogma?
1. DNA (Deoxyribonucleic Acid): Stores genetic information.
- Role: Template for transcription.
2. RNA (Ribonucleic Acid)
There are several types:
- mRNA (messenger RNA): Carries the code from DNA to ribosomes.
- tRNA (transfer RNA): Brings amino acids to the ribosome.
- rRNA (ribosomal RNA): Structural and enzymatic part of the ribosome.
3. Proteins (Polypeptides): Perform cellular functions (enzymes, structural proteins, signaling, etc.).
- Role: Final product of gene expression.
4. RNA Polymerase: Enzyme that synthesizes RNA from the DNA template during transcription.
5. Ribosome: Site of translation, where mRNA is decoded to build a protein.
6. Amino Acids: Building blocks of proteins.
Role: Assembled into a polypeptide chain based on the mRNA sequence.
7. Codons: Triplets of nucleotides in mRNA that specify amino acids.
Example: AUG = start codon (methionine)
8. Genetic Code: Matches mRNA codons with amino acids.
- Note: Universal and redundant (multiple codons can code for the same amino acid).
- Reverse Transcriptase (in viruses like HIV): Makes DNA from RNA, reversing the usual direction.
• Slide 7—is a great way to make sure you know all the information for this exam on transcription and translation.
•What regulates gene expression?
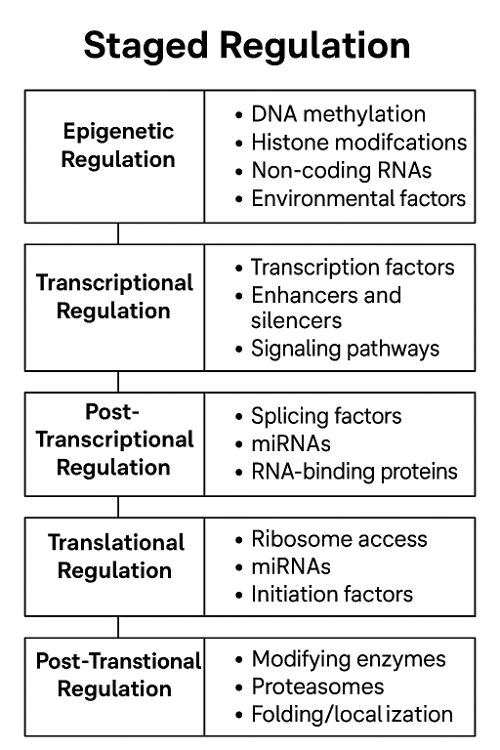
Gene expression is regulated at multiple levels by molecular mechanisms that control when, where, and how much of a gene's product is made. Key regulators include:
1. Epigenetic Regulation
- DNA methylation: addition of methyl groups to DNA (usually silences genes).
- Histone modification: alters how tightly DNA is wrapped around histones, affecting accessibility.
2. Transcriptional Regulation
- Transcription factors: proteins that bind DNA at promoters or enhancers to activate or repress transcription.
- Promoters: DNA sequences near the gene that help initiate transcription.
- Enhancers/silencers: DNA regions that increase or decrease transcription from a distance.
3. Post-Transcriptional Regulation
- Alternative splicing: different mRNA variants from the same gene.
- mRNA stability: how long mRNA lasts before it's degraded.
- microRNAs (miRNAs): small RNAs that bind to mRNA and block translation or cause degradation.
4. Translational Regulation
- Ribosome binding control: some sequences or proteins prevent ribosomes from attaching to mRNA.
- Regulatory proteins or miRNAs can also inhibit translation directly.
5. Post-Translational Regulation
- Protein modifications: phosphorylation, ubiquitination, etc., can activate or deactivate proteins.
- Protein degradation: tagging proteins with ubiquitin marks them for destruction.
•How does positive and negative regulation work?
Positive Regulation: A regulatory protein activates gene expression.
How it works:
- A transcriptional activator binds to DNA (e.g., enhancer or promoter region) and increases the rate of transcription.
- Example: The CAP protein in E. coli binds near the lac operon and activates transcription when glucose is low.
Analogy: Like pressing the gas pedal—you help turn the gene "on" or turn it on more strongly.
Negative Regulation: A regulatory protein inhibits gene expression.
How it works:
- A repressor binds to the DNA (often at an operator region) and blocks RNA polymerase from transcribing the gene.
- Example: The lac repressor binds to the lac operon and prevents transcription unless lactose is present.
Analogy: Like pressing the brakes—you stop the gene from being expressed.
•Describe the difference between operons and complex transcriptional activation.
Operons (Mostly in Prokaryotes like E. coli): A group of genes transcribed together from a single promoter into one mRNA.
Purpose: Allows coordinated expression of related genes.
Structure:
- Promoter – where RNA polymerase binds
- Operator – where a repressor/activator binds
- Genes – multiple coding sequences (often functionally related)
Example: Lac operon (controls lactose metabolism in E. coli)
Simple, efficient on/off switch for multiple genes in prokaryotes.
Complex Transcriptional Activation (Mostly in Eukaryotes): Involves multiple regulatory elements and transcription factors to control one gene.
Features:
- Enhancers and silencers (can be far from the gene)
- Combinatorial control – many transcription factors must bind together
- Chromatin remodeling – accessibility of DNA is regulated
Purpose: Allows fine-tuned, tissue-specific, and time-specific gene expression.
Example: Activation of the β-globin gene in red blood cell precursors.
Sophisticated, flexible control used in eukaryotes, often for just one gene at a time.
•How does the lac operon work? How does the tryptophan operon work? Are they positive or negative regulators?
Lac Operon (Lactose Operon)
Goal: Break down lactose.
- Normally off
- Turns on when lactose is present
- Lactose removes a repressor (a protein that blocks the gene) → gene turns on
- Works even better when glucose is low (a helper protein called CAP boosts it)
Uses both: Negative regulation (repressor blocks it) and Positive regulation (CAP helps turn it on)
Trp Operon (Tryptophan Operon)
Goal: Make tryptophan.
- Normally on
- Turns off when tryptophan is present
- Tryptophan helps a repressor attach → gene shuts off
Uses only: Negative regulation (repressor stops it when enough tryptophan is made)
•How does epigenetics play a role in regulating gene expression?
Epigenetics controls gene activity without changing the DNA sequence.
Key ways it does this:
- DNA methylation: adds methyl groups to DNA → often silences genes
- Histone modification: changes how tightly DNA is wrapped around histones
- Tightly wrapped = genes off
- Loosely wrapped = genes on
- These changes are reversible and can be passed to daughter cells
Think of it as controlling gene access like a light dimmer, not an on/off switch.
•What is combinatorial control?
Combinatorial control means a gene is turned on/off by many transcription factors working together.
A single gene may need:
- An activator to start transcription
- One or more co-activators
- No repressors
- Specific timing, location, or signals
Like using multiple keys to unlock one door—specific combinations control expression.
•What regulates transcription factors?
- Cell signals (like hormones or growth factors)
- Phosphorylation (adds a phosphate group → turns them on/off)
- Binding to other proteins
- Localization (they may need to enter the nucleus to work)
What regulates activators?
- Ligand binding (e.g., steroid hormones activate their receptors)
- Post-translational modifications (like phosphorylation)
- Presence of co-activators or helper proteins
- Epigenetic state of the DNA (if DNA is too tightly packed, even activators can’t help)
What regulates repressors?
- Corepressors (molecules that help them bind DNA)
- Signals that remove or block them
- Allosteric changes (shape changes caused by binding a molecule)
- Proteolysis (targeted destruction of the repressor protein)
What regulates staged regulation?
•What are the 3 main steps of transcription?
1. Initiation: RNA polymerase attaches to the DNA at the start of the gene (promoter).
- DNA unwinds so it can be read.
- Starting the copy.
2. Elongation: RNA polymerase builds an RNA strand using one DNA strand as a guide.
- It adds A, U, C, and G bases to make the RNA.
- Copying the message.
3. Termination: RNA polymerase reaches the end of the gene.
- RNA is released, and the DNA rewinds.
- Stopping the copy.
•What is main differences between prokaryotic and eukaryotic transcription?
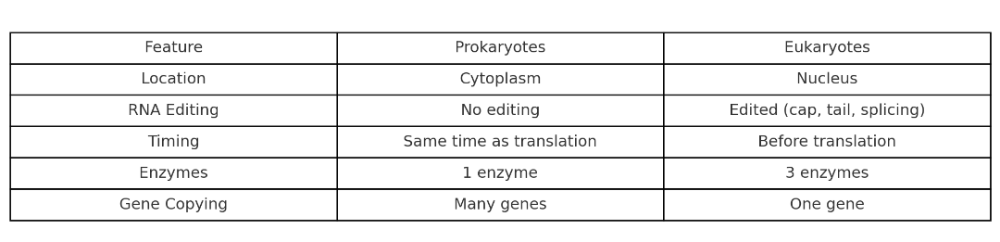
Where does transcription happen in prokaryotes vs eukaryotes?
- Prokaryotes: In the cytoplasm (they don’t have a nucleus)
- Eukaryotes: In the nucleus (then mRNA moves to the cytoplasm for translation)
Key point: Only eukaryotes separate transcription and translation by location.
What are the main proteins in transcription of prokaryotes and eukaryotes?
Prokaryotes
Main proteins:
1. RNA Polymerase
- One main enzyme does all transcription
- Has a core enzyme and a sigma factor
- Sigma factor helps it find the promoter
2. Sigma Factor
- Guides RNA polymerase to the correct DNA sequence (promoter)
Simple setup: One polymerase, one helper.
Eukaryotes
Main proteins:
1. RNA Polymerases I, II, III
- RNA Pol II transcribes most protein-coding genes
- Pol I → rRNA, Pol III → tRNA and small RNAs
2. General Transcription Factors (GTFs)
- Examples: TFIID, TFIIB, TFIIH
- Help RNA Pol II bind to the promoter and start transcription
3. Mediator Complex
- Helps connect transcription factors and RNA Pol II
- Acts as a “bridge” for regulation and coordination
More complex: Multiple polymerases + many helper proteins
Does splicing happen in the transcription of prokaryotes and eukaryotes?
Prokaryotes
- No splicing
- Their genes don’t have introns, so nothing needs to be cut out.
Eukaryotes
- Yes, splicing happens
- Eukaryotic genes have introns (non-coding parts)
- Splicing removes introns and joins exons
- Happens after transcription, but before translation
Only eukaryotes need to edit the RNA before using it.
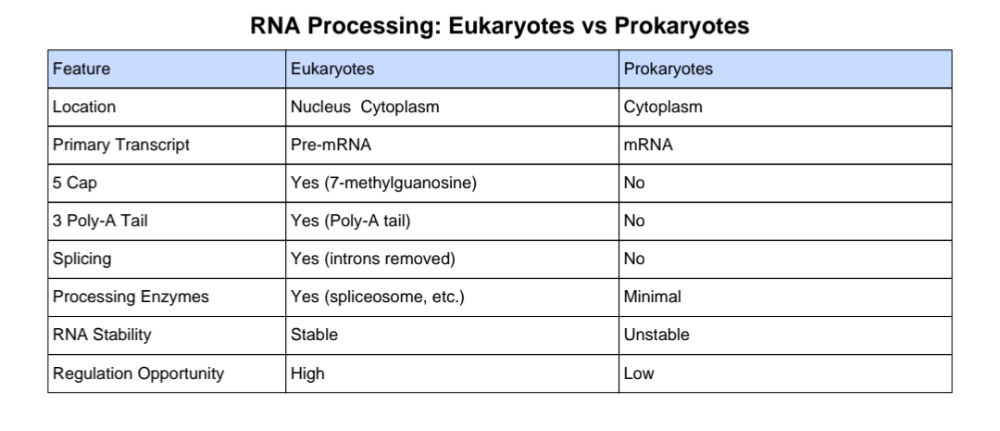
What are the differences between RNA processing in eukaryotes and prokaryotes?
•What does RNA Polymerase I transcribe? What other transcription factors help? Describe the process.
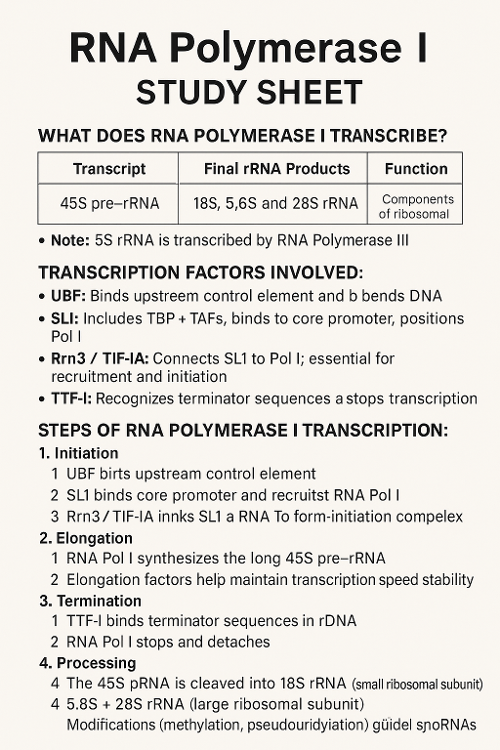
•What does RNA Polymerase III transcribe? What other transcription factors help? Describe the process.
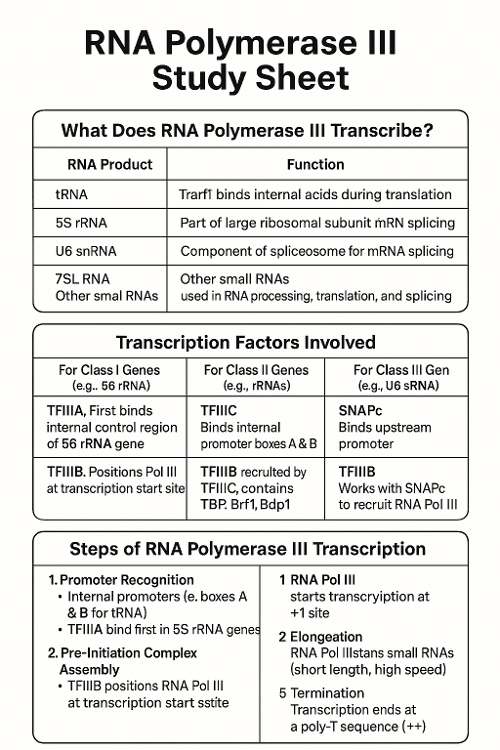
•What does RNA Polymerase II transcribe? What other transcription factors help? Describe the process.
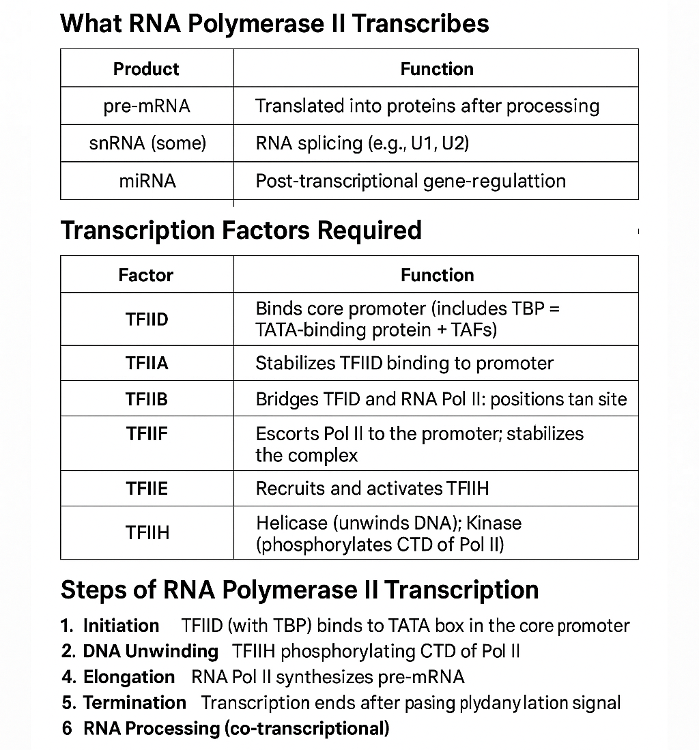
•Describe the steps of mRNA processing

•What goes on the 5’ end of mRNA? The 3’ end? Why?
5′ End of mRNA
- Modification: 7-methylguanosine cap (also called the 5′ cap)
- Attached: Shortly after transcription begins, via an unusual 5′–5′ triphosphate linkage
Functions:
- Protects mRNA from exonuclease degradation
- Helps with nuclear export
- Aids ribosome recognition and binding for translation initiation
3′ End of mRNA
- Modification: Poly-A tail (a string of ~150–250 adenine nucleotides)
- Added: After cleavage at the polyadenylation signal (AAUAAA)
Functions:
- Increases mRNA stability
- Aids in nuclear export
- Enhances translation efficiency
- Helps regulate mRNA lifespan
•Describe the spliceosome and the spliceosome machinery?
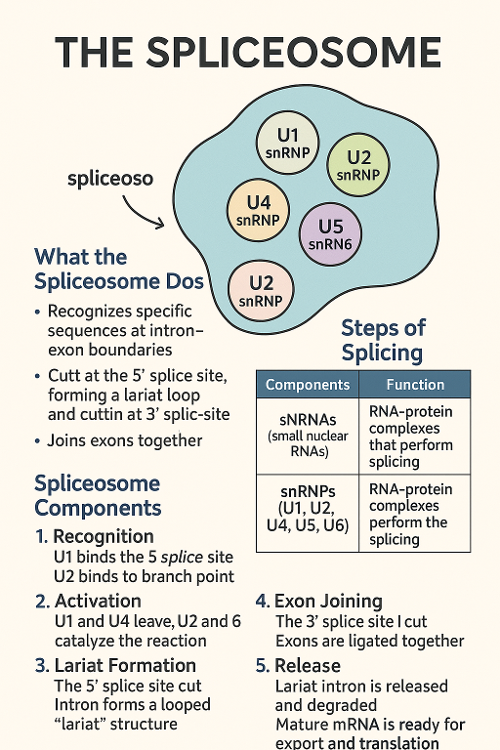
•What is alternative splicing?
when a single gene can make different versions of mRNA, depending on how the exons are put together. This lets one gene make different proteins.
What Happens:
Exons = coding parts of a gene
The spliceosome can:
- Keep some exons
- Skip others
- This changes the final mRNA and protein.
Why It Matters
- More proteins: One gene can make many types of proteins
- Cell control: Different cells can make different protein versions
- Adaptation Cells: can change proteins based on signals
•How does mRNA exit the nucleus?
1. mRNA Processing Completed
- Only fully processed (mature) mRNA is allowed to leave.
- Must have: 5′ cap, Spliced exons, and Poly-A tail
2. Binding of Export Proteins
- Special proteins bind to the mRNA, recognizing it as ready.
- Key complex: TREX (Transcription-Export) complex
- Cap-binding proteins and exon junction complexes also help guide export.
3. Transport Through the Nuclear Pore Complex (NPC)
- mRNA exits through nuclear pore complexes, large gateways in the nuclear envelope.
- Transport is energy-dependent and selective
4. Release into the Cytoplasm
- Export proteins are removed.
- mRNA is handed off to ribosomes for translation.
•What are the 4 ways mRNA is degraded?
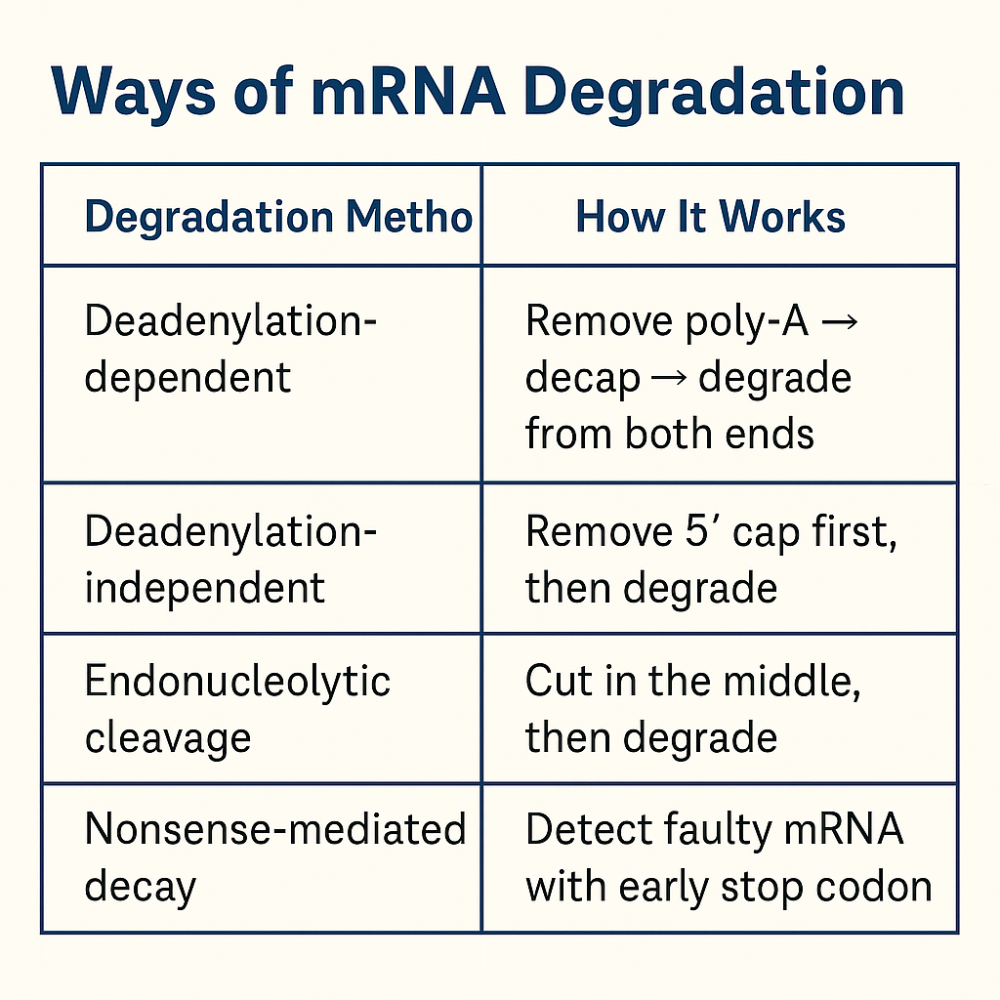
•Describe the steps of tRNA processing
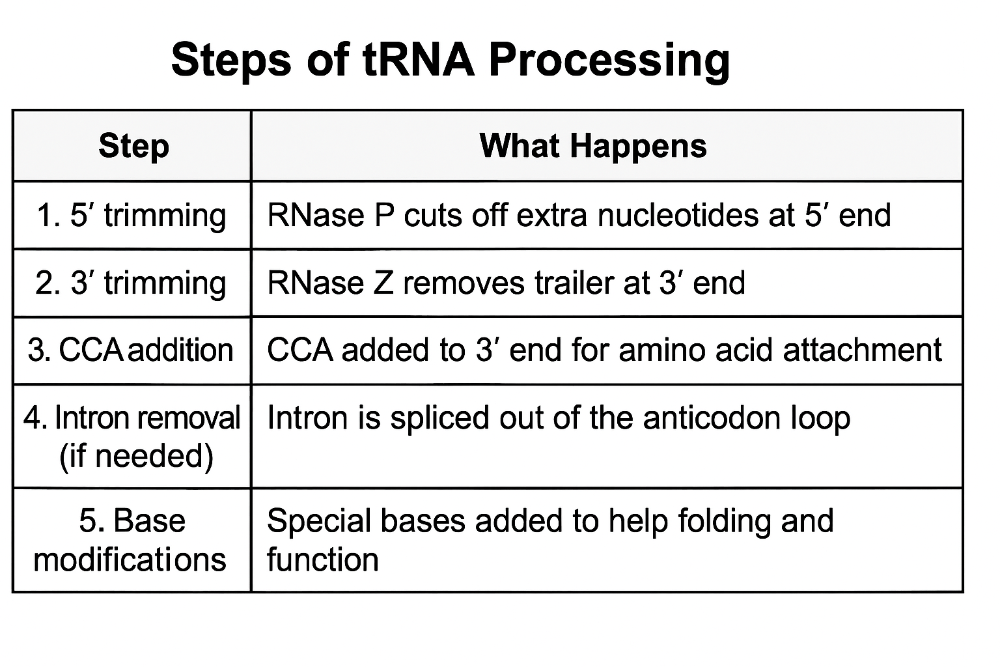
•Describe the steps of rRNA processing
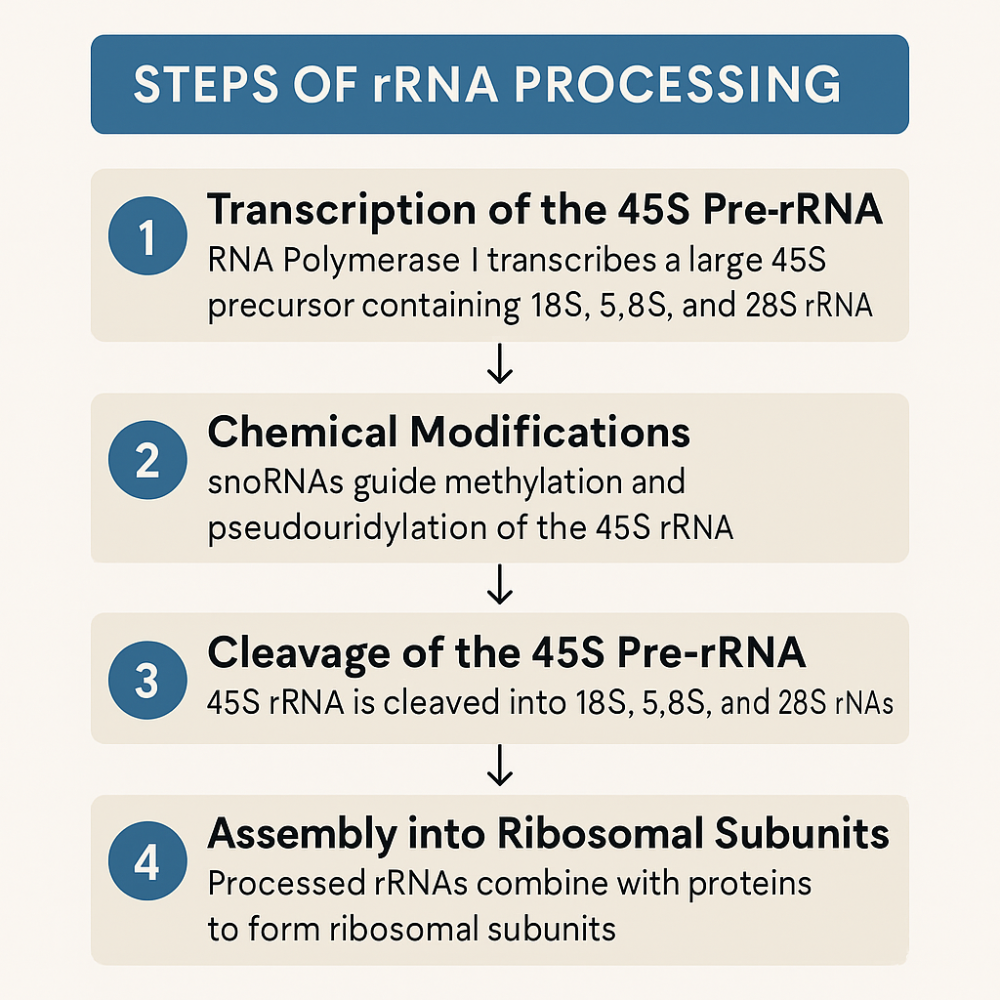
•What is snoRNA?
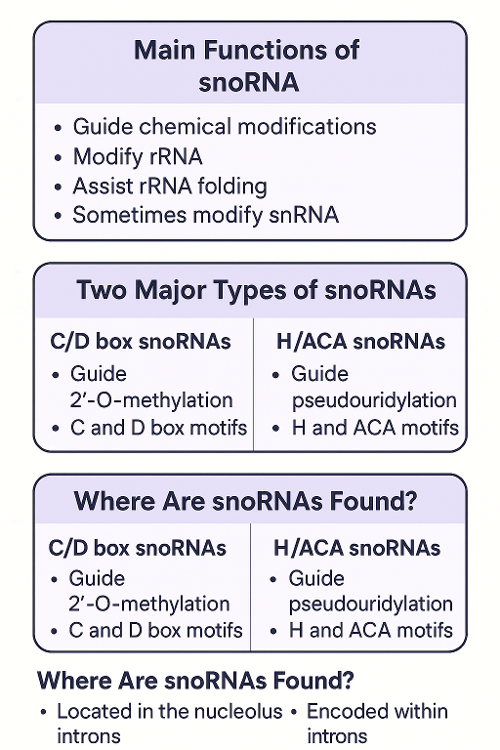
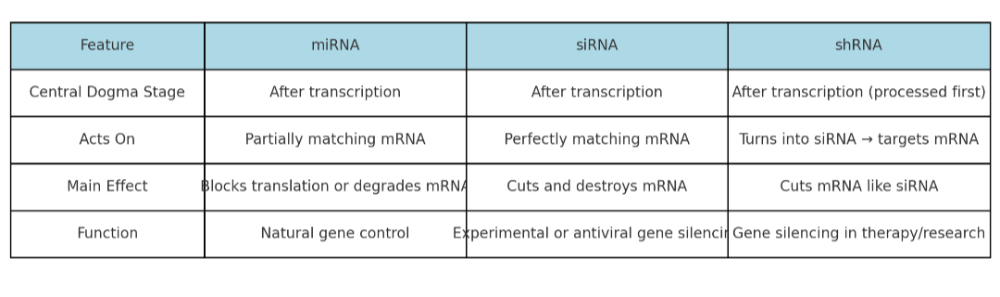
•Describe the role of siRNA, miRNA, and shRNA. How are they the same? Different?
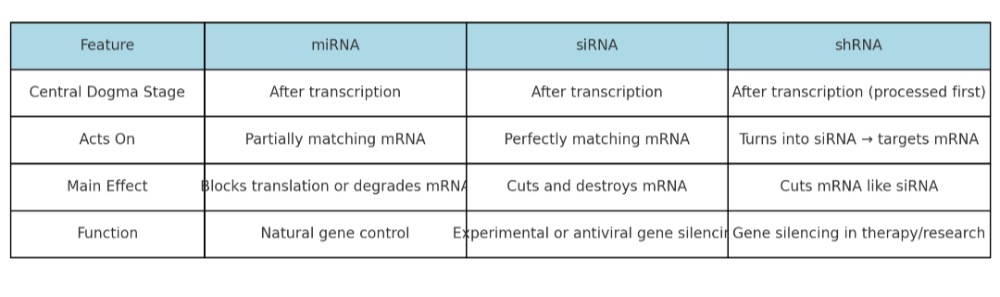
•Integrate the roles of the different types of RNA in the central dogma
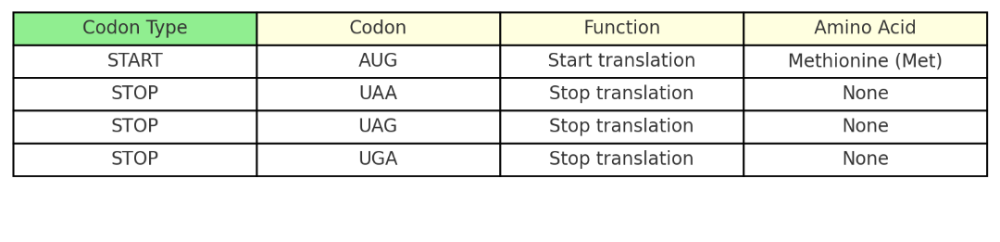
What are START and STOP codons?
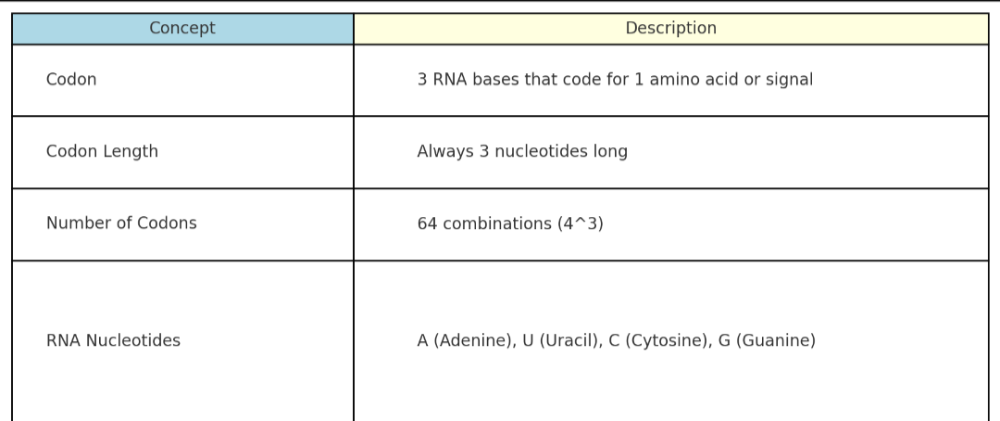
•What are codons? How many nucleic acids are in RNA?
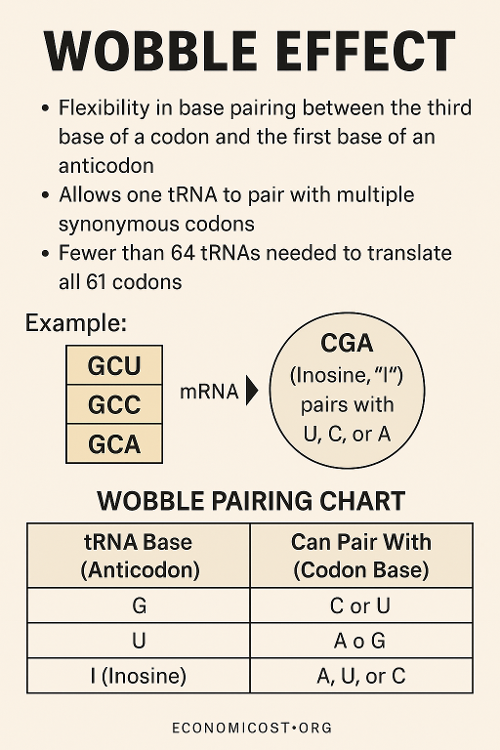
•What is the wobble effect?
•Why and how is tRNA charged?
Why is tRNA Charged?
- Link between mRNA and protein: Charged tRNA brings the right amino acid to the ribosome
- Translation accuracy: Ensures the codon matches the correct amino acid
- Required for peptide bond formation: Ribosome needs aminoacyl-tRNA to grow the polypeptide chain
How is tRNA Charged?
1. Activation of Amino Acid
- Amino acid + ATP → Aminoacyl-AMP + PPi
- Forms a high-energy intermediate
2. Transfer to tRNA
- Aminoacyl group is transferred to the 3′ end (CCA tail) of the tRNA
- Forms aminoacyl-tRNA
Result:
- A charged tRNA (also called aminoacyl-tRNA)
- Ready to deliver its amino acid during translation
•How are ribosomes formed?
1. rRNA Transcription (in Nucleolus)
- 45S pre-rRNA → cleaved into 18S, 5.8S, and 28S rRNAs
- 5S rRNA is made separately in the nucleoplasm
2. Ribosomal Protein Import
- Ribosomal proteins are made in the cytoplasm from mRNA
- They are imported into the nucleus, then the nucleolus
3. Pre-ribosome Assembly
rRNAs and proteins come together to form:
- Pre-40S subunit (with 18S rRNA)
- Pre-60S subunit (with 5.8S, 28S, and 5S rRNA)
4. Processing & Maturation
- More modifications, cleavages, and folding steps
- Involves helper proteins and snoRNAs
5. Export to Cytoplasm
- The pre-40S and pre-60S subunits are transported through nuclear pores
- Once in the cytoplasm, final maturation occurs
- The two subunits join during translation initiation
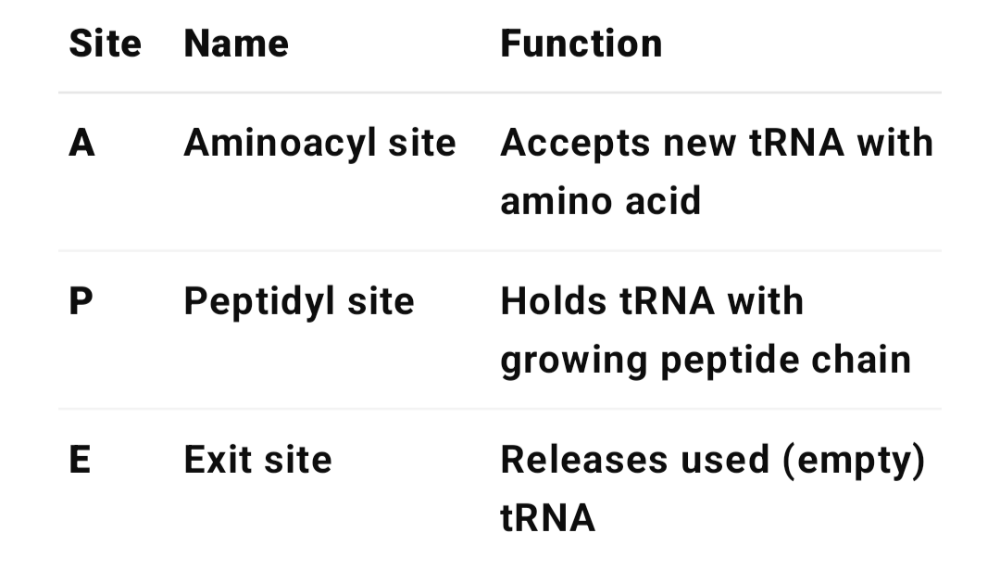
•What is the purpose of the E, P and A sites?
•What is the purpose of a poly ribosome?
a cluster of multiple ribosomes attached to a single mRNA strand, all translating the mRNA at the same time.
Purpose of a Polyribosome:
- Increases efficiency: Many ribosomes make many copies of the same protein at once
- Speeds up protein production: No need to wait for one ribosome to finish before starting
- Conserves resources: One mRNA produces multiple proteins before degrading
- Supports high-demand needs: Critical for fast-growing or protein-producing cells (like muscle or liver cells)
•How does the rough ER form?
1. Starts as Smooth ER: The ER begins as a network of membranes (smooth ER) extending from the nuclear envelope.
2. Ribosomes Bind to the ER Membrane: Ribosomes making secreted or membrane-bound proteins attach to the ER membrane. This happens through a signal peptide on the growing protein that is recognized by the signal recognition particle (SRP). SRP directs the ribosome to the ER membrane, where it docks at a translocon (protein channel).
3. Ribosomes Stay Attached During Translation
As the protein is synthesized, it is inserted directly into the ER lumen or membrane. This ribosome attachment gives the ER a "rough" appearance.
4. Rough ER Expands with Protein Production
The more ribosomes bind for protein synthesis, the more extensive the rough ER becomes. The membrane system grows and is maintained by the cell’s need for protein processing and trafficking.
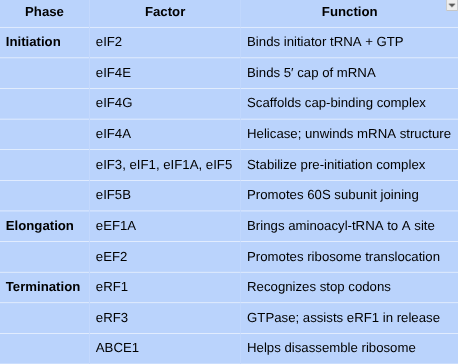
•What are the main steps in translation?

•Describe the steps of translation, know the list of proteins/translation factors associated with translation
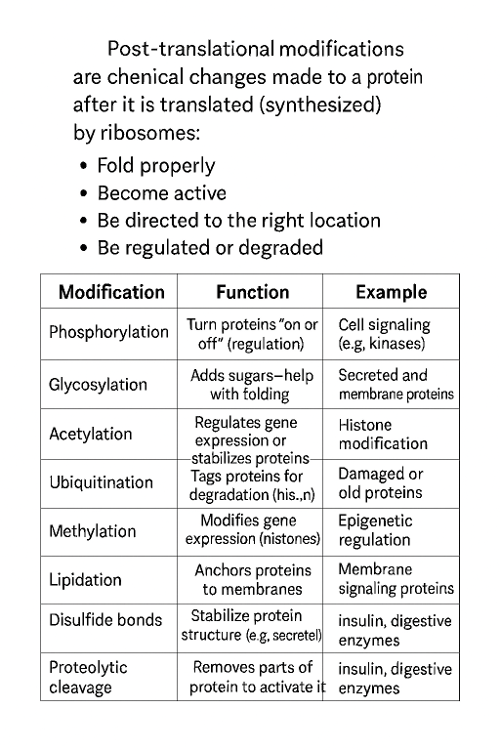
•What are post translational modifications?
•What is the purpose of a chaperone protein?

•What happens when chaperone proteins do not work?
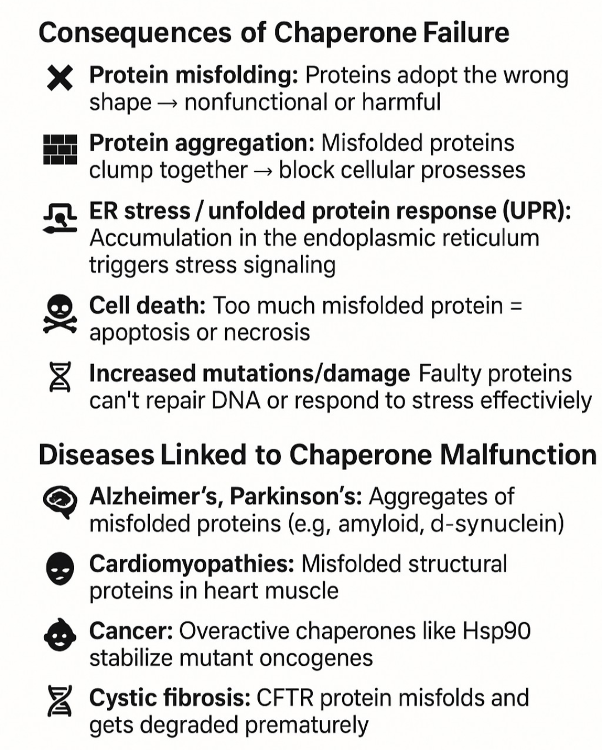
Prion diseases: Misfolded prion proteins (e.g., PrPᶜ → PrPˢᶜ) cause a chain reaction of misfolding
•Why is protein folding important? What bonds help with this?
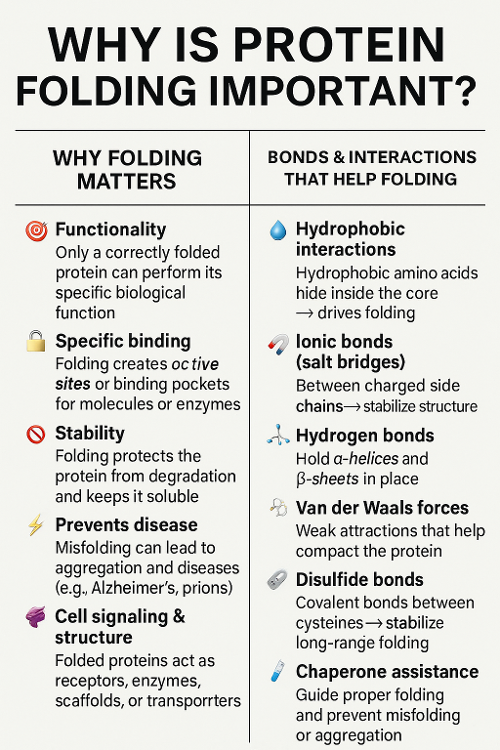
•Why is cleavage of pieces of proteins important?

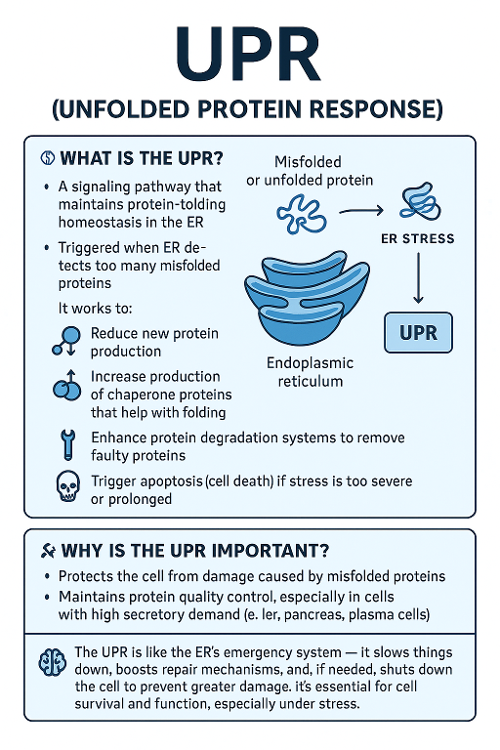
•What is the UPR? Why is it important?
•List specific post translational modifications.
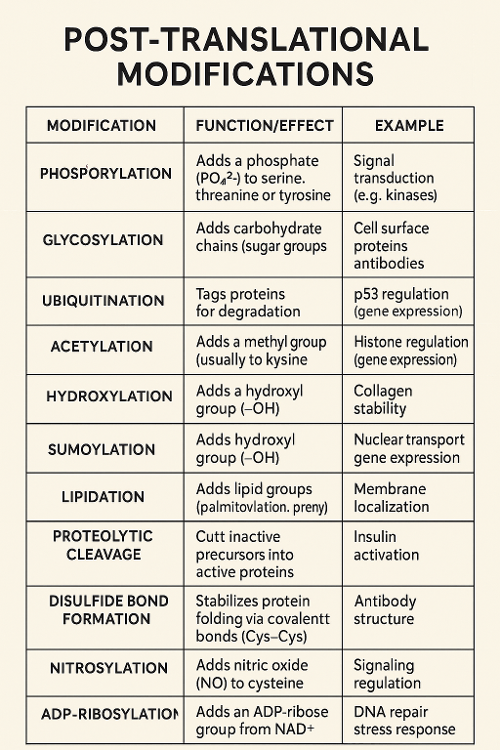
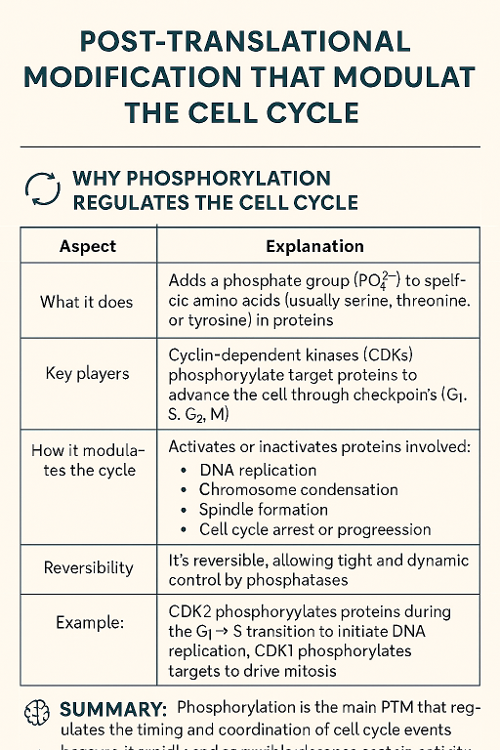
•Tie back to previous chapter: Which post translational modification modulates cell cycle? Why?
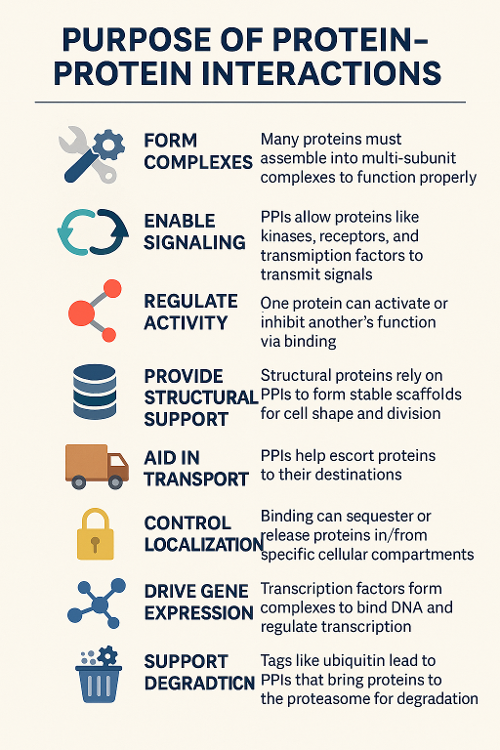
•What is the purpose of protein protein interactions?
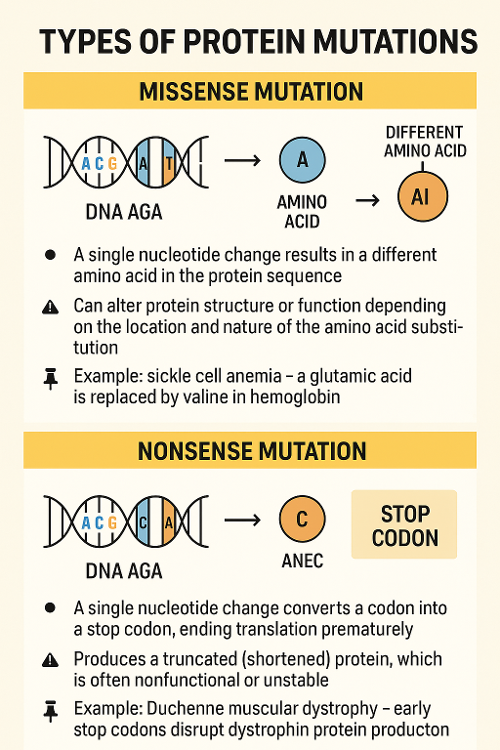
•Describe two kinds of protein mutations?
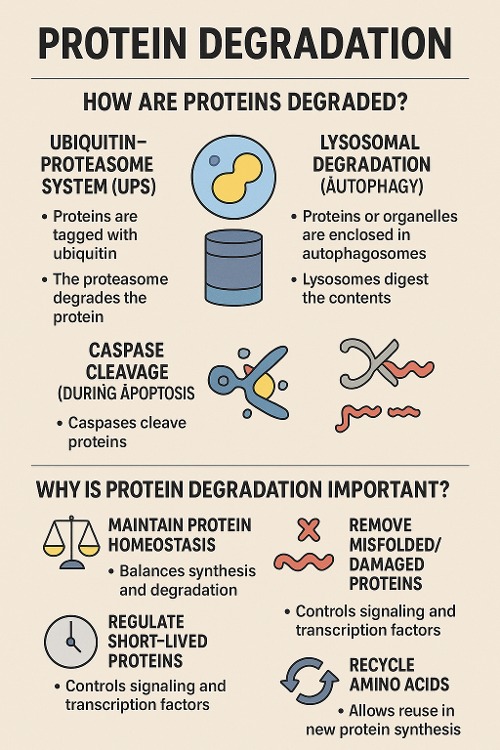
•How are proteins degraded? Why is that important?
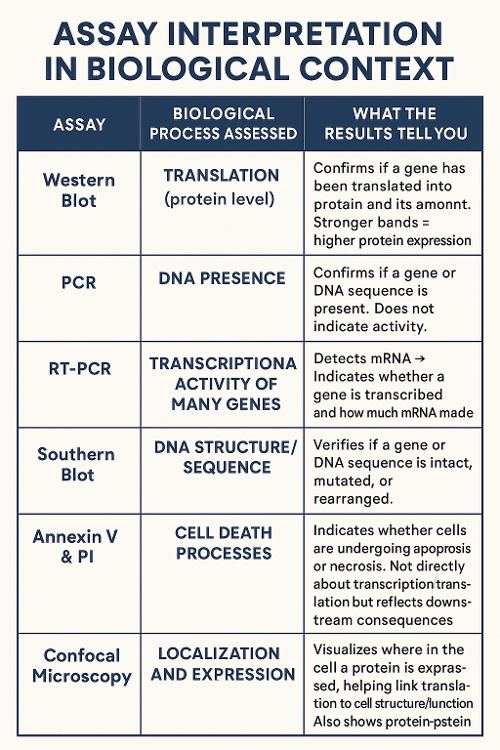
•What do each of the following assays measure and how can you read the results?
Western Blot, PCR, Reverse Transcriptase PCR, Microarrayo Southern Blot, Annexin V and Propidium Iodide and Confocal Microscopy
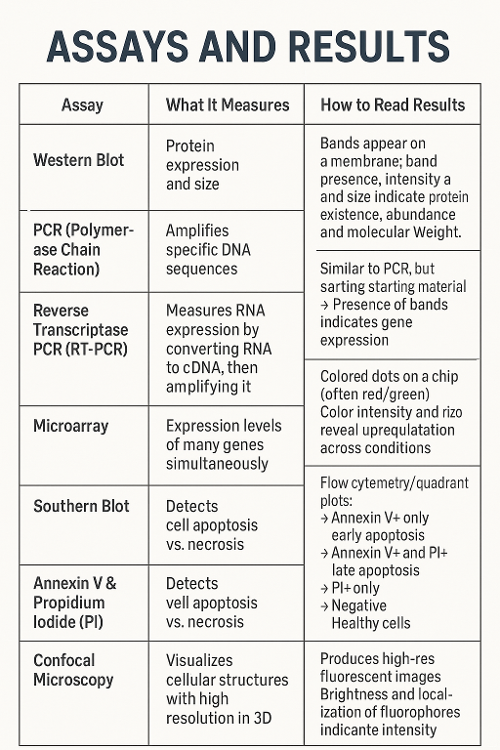
•What do the results mean in response to transcription and translation? Or another process
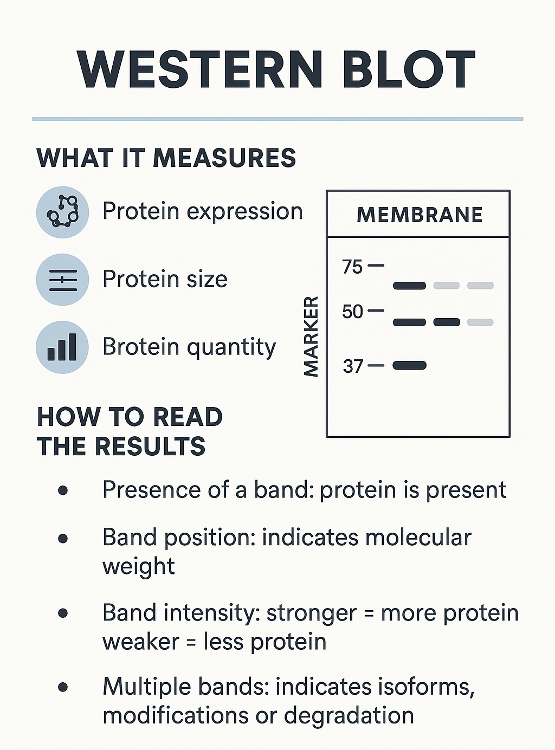
Western Blot
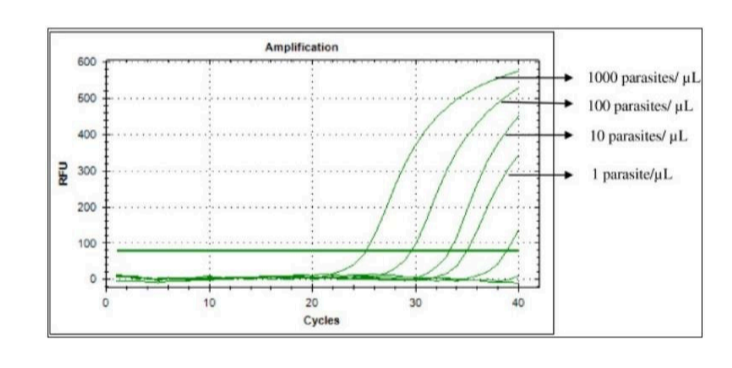
PCR

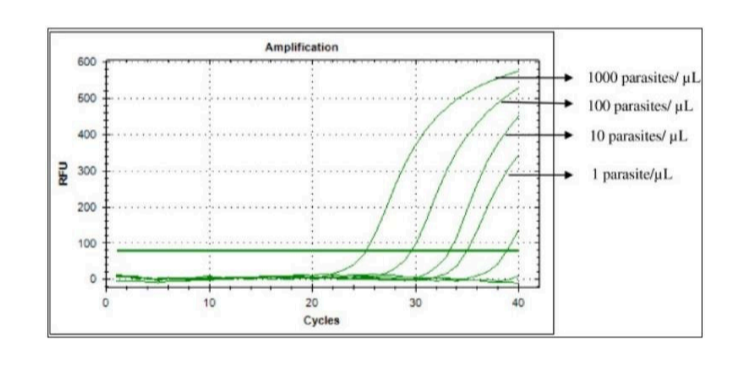
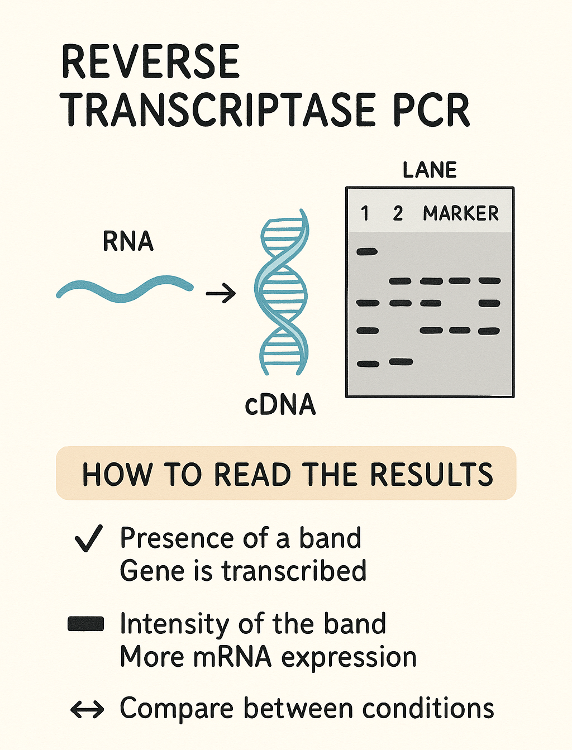
Reverse transcriptase PCR

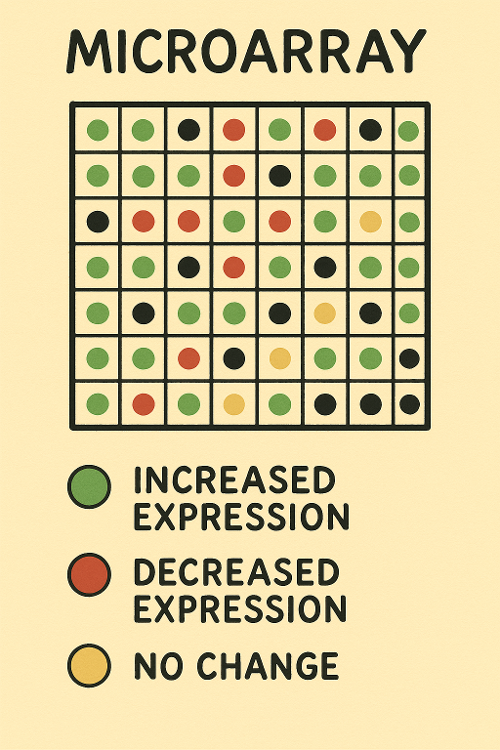
Microarray
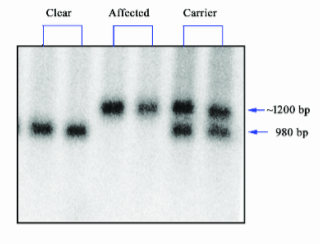
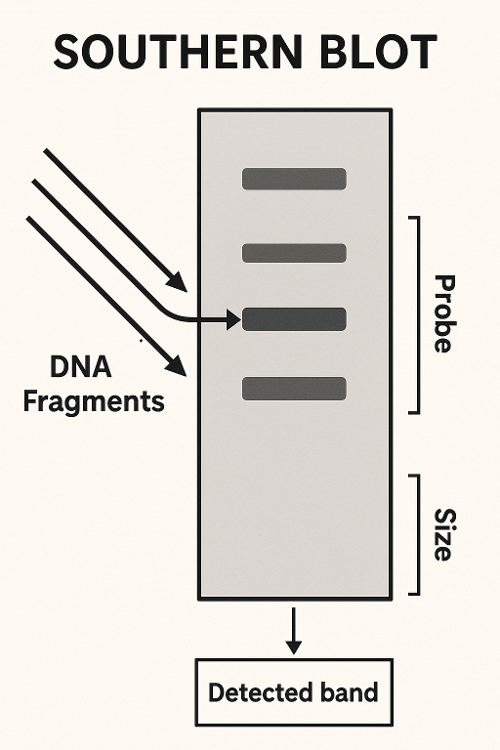
Southern Blot
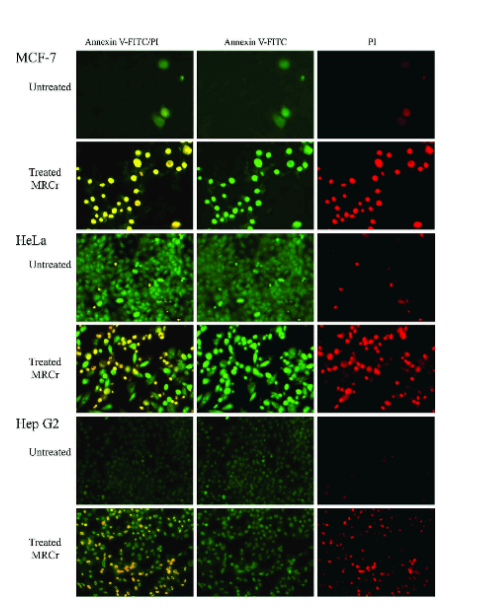
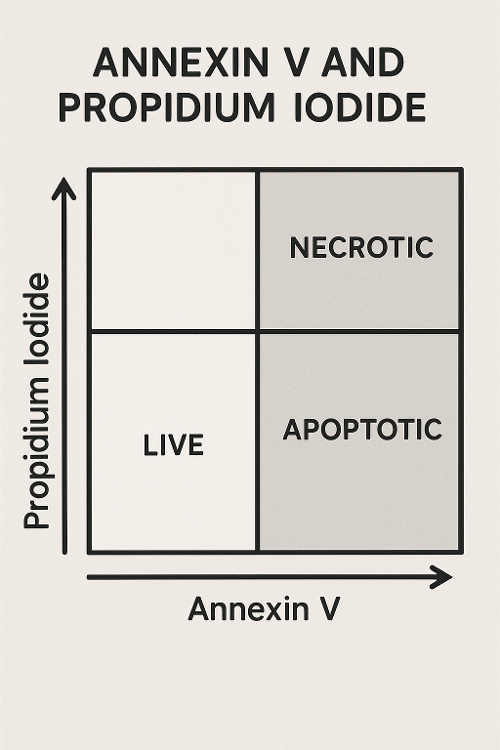
Annexin V and Propidium Iodide
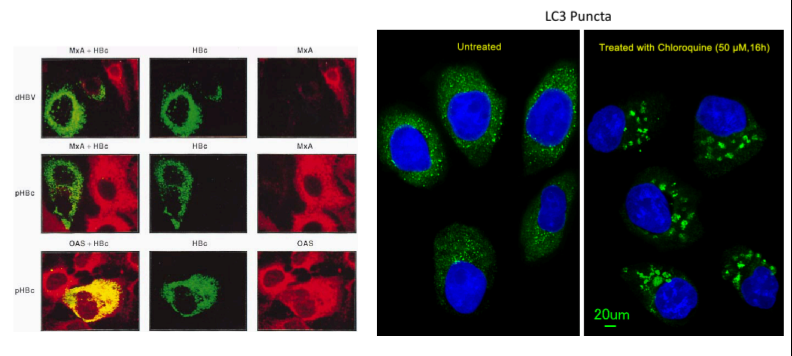
Confocal Microscopy

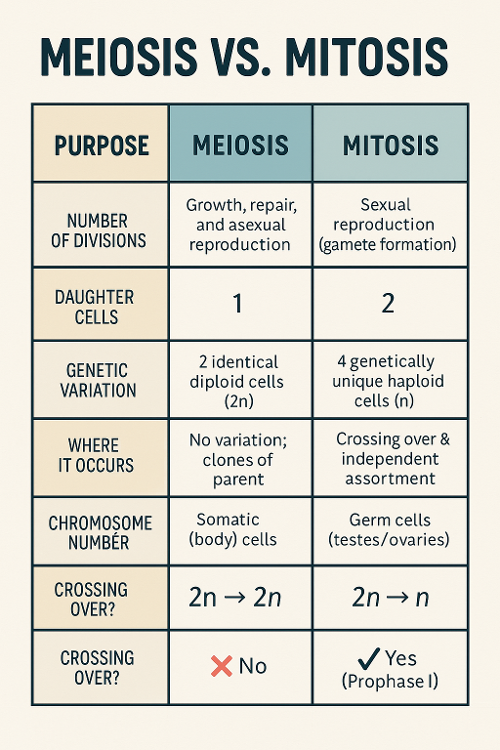
•Describe the difference between meiosis and mitosis.
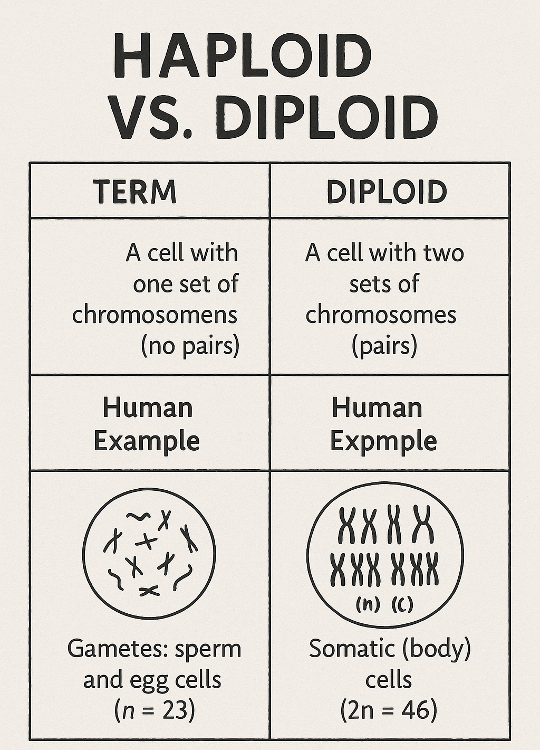
•What is the difference between haploid and diploid? What are the number of chromosomes in a human?
•What are the two ways to increase genetic diversity in meiosis?
1. Crossing Over (Recombination)
- Occurs in: Prophase I of meiosis
- What happens: Homologous chromosomes exchange genetic material at chiasmata.
- Result: New combinations of alleles on each chromosome — increases variation in offspring.
2. Independent Assortment
- Occurs in: Metaphase I of meiosis
- What happens: Homologous chromosome pairs line up randomly at the cell’s equator.
- Result: Each gamete gets a random mix of maternal and paternal chromosomes.
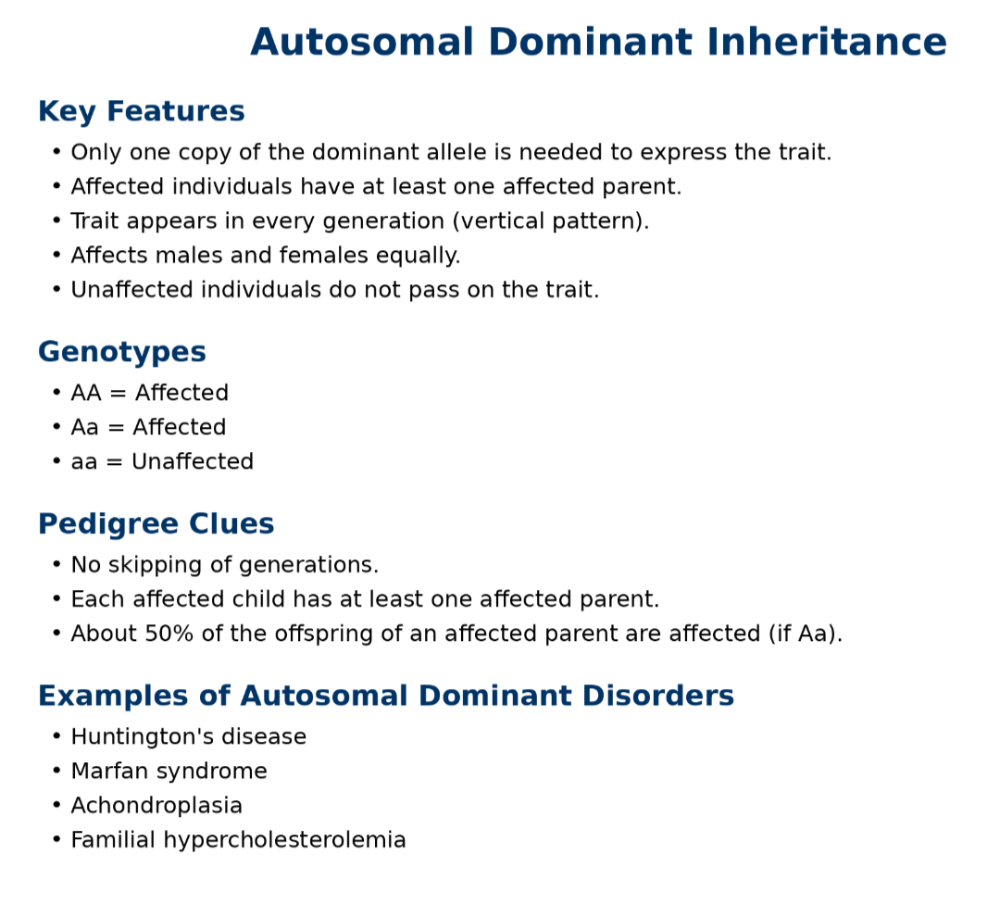
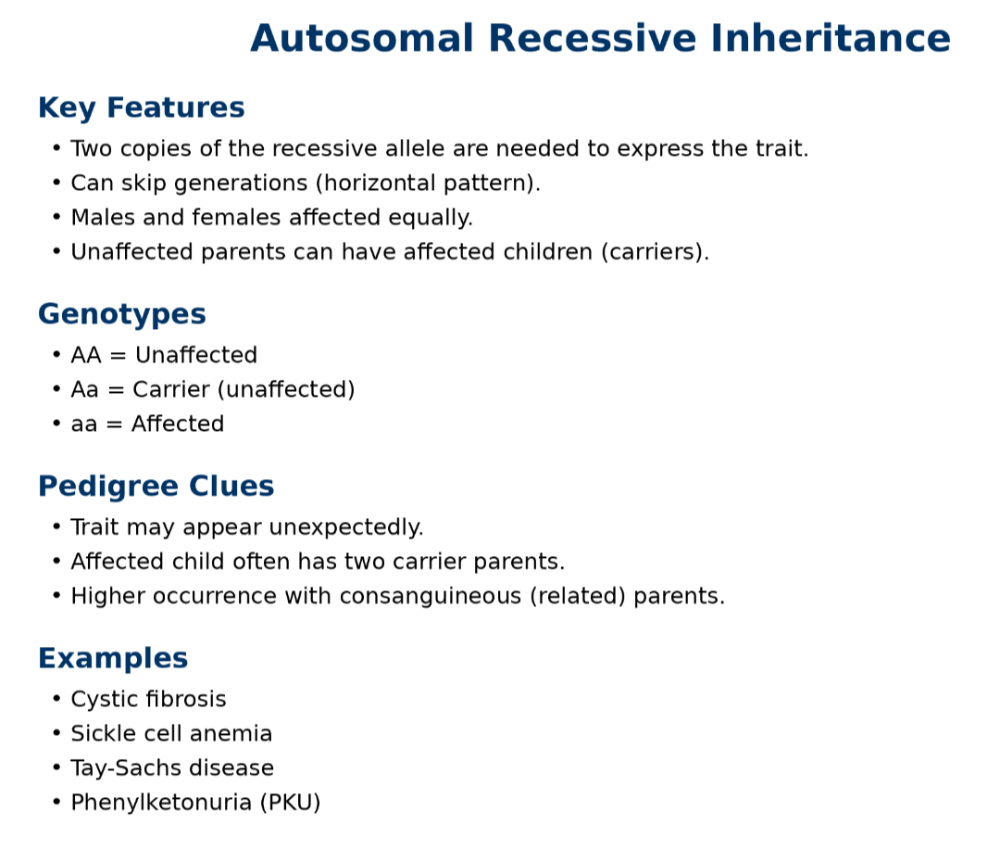
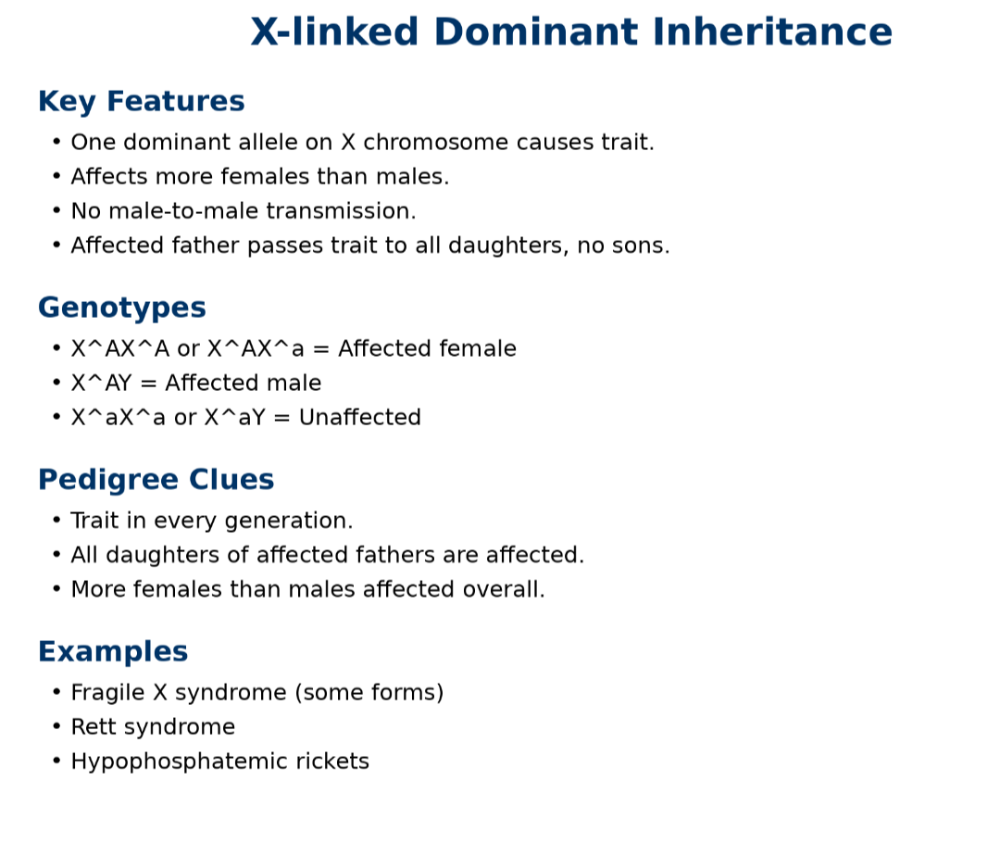
•Understand dominant and recessive traits

•Be able to read pedigree trees
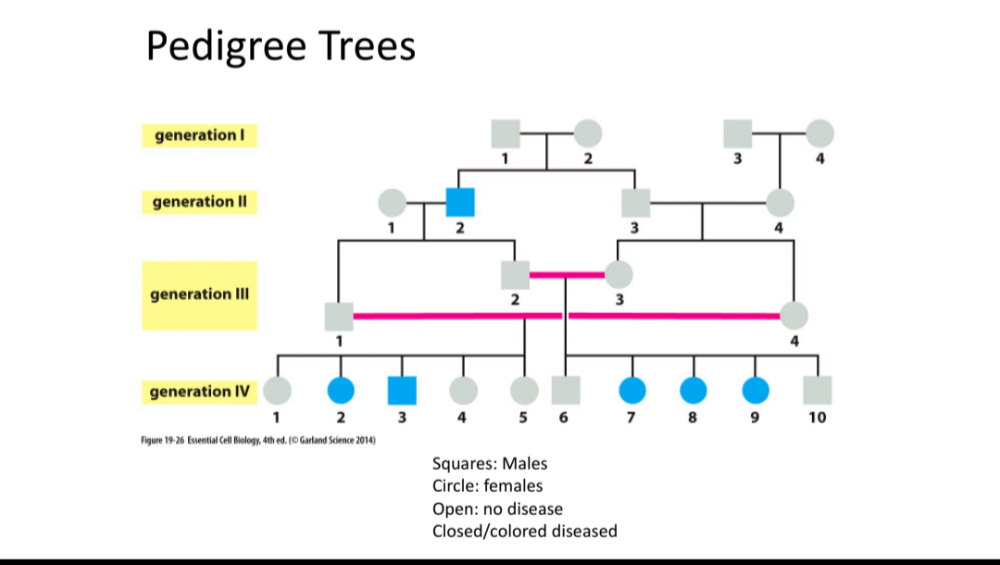
oAutosomal dominant
oAutosomal recessive
oX linked dominant
oX linked recessive

Y linked

Which of the following proteins contains helicase activity?
A. TFIIH
B. TFIIE
C. TFIIIH
D. TFIIIE
A. TFIIH
Which of the following polymerase transcribes most rRNA?
A. RNA Polymerase I
B. RNA Polymerase II
C. DNA Polymerase III
D. RNA Polymerase III
A. RNA Polymerase I
Which of the following factors stops repressors from binding to DNA?
A. TFIIA
B. TFIID
C. TFIIB
D. TFIIE
A. TFIIA
Which of the following factors binds to the TATA box on the DNA?
A. TFIIA
B. TFIID
C. TFIIB
D. TFIIE
B. TFIID
Which Transcription factor regulates CTD on RNA Polymerase II?
A. TFIIG
B. TFIIB
C. TFIIH
D. TFIIA
C. TFIIH
Which translation factor binds to the 5' cap of mRNA?
A. eIF1
B. eIF2
C. eIF3
D. eIF4
B. elF2
Where does the first tRNA of translation bind?
A. AUG in the P site
B. AUG in the E site
C. AUG in the A site
D. Anywhere in the molecule that matches the codon
A. AUG in the P site
Which translation factor is essential for the cleavage of GTP to allow the large ribosome to join translation?
A. eIF2
B. eIF1
C. eIF4
D. eIF5
D. eIF5
Which site does the amino acid chain bind?
A) A
B) E
C) P
A) A
Which translation factor binds to UAG?
A. eRF1
B. eEF1
C. eIF2
D. eEF2
A. eRF1
Which factor is essential for the creation of the Rough ER?
A. TFIID
B. Sec61
C. eRF1
D. eIF4
B. Sec61
What does RT (Reverse Transcriptase) PCR measure?
A. Transcription
B. Both
C. Translation
D. Neither
A. Transcription
What does PCR measure?
A. Transcription
B. Both
C. Translation
D. Neither
D. Neither
What does Microarray measure?
A. Transcription
B. Both
C. Translation
D. Neither
A. Transcription
What does Southern Blot measure?
A. Transcription
B. Both
C. Translation
D. Neither
D. Neither
What does Western Blot measure?
A. Transcription
B. Both
C. Translation
D. Neither
C. Translation
To access, central Dogma what do you want to study?
A. Transcription
B. Both
C. Translation
D. Neither
B. Both