biochem cell bio exam 1 10/1
difference between covalent and non covalent bonds
atoms in a covalent bond share some of their electrons, but the atoms in a non-covalent bond do not share any electrons.
Which term describes all types of noncovalent bonds
electrostatic
London dispersion forces are attractive forces that arise when ________
oppositely charged dipoles, created through the random distribution of electrons around their atoms’ nuclei, interact
Which statements are TRUE (select all)?
A) Dipole-dipole interactions must involve a hydrogen
atom.
B) Hydrogen bond formation is possible between -CH and-NH3
.
C) The carboxylic acid (COOH) functional group could form
hydrogen bonds with water.
D) An ionic bond could form between
two carboxylate (COO- ) functional groups.
E) Hydrocarbon chains
can interact through dispersion forces.
The carboxylic acid (COOH) functional group could form hydrogen bond with water
Hydrocarbon chains can interact through dispersion forces
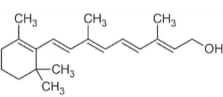
The molecule shown below is primarily ______ because ___________
non-polar; it mostly contains atoms with similar electronegativity properties
Which molecule below is the most hydrophilic (water loving)? Hint: This molecule will be highly polar
Urea
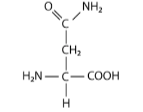
This amino acid below is in the _____ category. It’s R-group has a _____ functional group, which means it will ______ when the pH is extremely low
Polar/amide/not change
hydroxyl
OH
Carboxylic acid
CO2H
Amine
NH2
Methyl
CH3
The primary structure of a protein is determined by the
the identity of the amino acids in the peptide sequence
Which is/are a true statement(s)? Use the figure and the table below
for reference
and select all correct answers.
A) The primary
amino acid sequence for myoglobin is identical to that
of
hemoglobin.
B) hemoglobin is tetrameric and has
quaternary structure.
C) myoglobin is monomeric and does not have
quaternary structure.
D) The structure of hemoglobin and
myoglobin mostly contain beta strands.
E) Hemoglobin binds to a
small molecule cofactor, but myoglobin doesn’t.
hemoglobin is tetrameric and has quaternary structure.
myoglobin
is monomeric and does not have quaternary structure.
Which of the following statements is TRUE (select all)?
A)
Peptide bonds are the only covalent bonds that can link together two
amino
acids in proteins.
B) The primary amino acid sequence
is always read from carboxy to amino
terminus
C) The
sequence of the atoms in the polypeptide backbone varies
between
different proteins.
D) The primary amino acid
sequence Proline-Alanine-Glycine-
Phenylalanine-Glycine is not
likely to occur in an alpha helix
The primary amino acid sequence
Proline-Alanine-Glycine-
Phenylalanine-Glycine is not likely to
occur in an alpha helix
A peptide bond is stronger than a single covalent bond because it has
a partial
double bond character. This means that not only is this
bond stronger than a single
bond, but it can also limit the
geometry of the molecule because (select one)
The rotation of the bond is limited due to resonance
Which statement correctly describes the bonds that form within a
protein? (select all)
A) Primary structure depends on hydrogen
bond formation between backbone
atoms.
B) Secondary
structure depends on ionic or dipole-dipole interactions
between
R-groups.
C) Tertiary structure only forms from
non-covalent interactions between R-
groups.
D) Quaternary
structure only involves interactions between multiple domains.
E)
None of the above
None of the above
Glycine and proline are known as “helix breakers”. Explain why
Both amino acids disrupt the regular hydrogen bonding pattern
essential for alpha
helical structure. Glycine’s R group is very
small and rotation around the phi and psi
bonds are quite
flexible. This flexibility makes it harder for the atoms to “settle”
into
the position conducive for H-bond formation. Proline’s
R-group is covalently bound
to the imide nitrogen, which means
that rotation in the phi-psi bond space is severely
restricted.
Consequently, proline is unable to complete the H-bonding chain of
the
helix and it will cause a “kink” in the alpha helical structure
Which of the following statements are true about PrPC and/or Prp Sc ?
(select one)
A. PrPC and Prp Sc have different primary structures
AND tertiary structures.
B. PrPc can spontaneously undergo the
conformational change to become PrPsc
C. Prions only infect
humans
D. Prp Sc causes cellular damage because it forms
aggregates through alpha helical
stacking
PrPc can spontaneously undergo the conformational change to become PrPsc
Protein folding is mostly driven by
hydrophobic effect
Molecular chaperones help the ∆F508 form of CFTR re-fold
False
Which of the following best describes the change in entropy that occurs during protein folding
A. Entropy of both the water and the protein increase.
B.
Entropy of the water increases; entropy of the protein
decreases.
C. Entropy of the water decreases; entropy of the
protein increases.
D. Entropy of both the water and the protein decrease
Entropy of the water increases; entropy of the protein decreases
The K D of Protein A for Ligand B is 300 nM. The KD of Protein C for
Ligand D is 10
μM. Which of the following statements are TRUE
(select all)?
A. These two dissociation constants cannot be
compared.
Week 3 Practice Problems | Biochem 285, FA24
Mearls
B. Protein A binds Ligand B with higher affinity than
Protein C binds Ligand D
C. Protein C binds Ligand D with higher
affinity than Protein A binds Ligand B
D. When the concentration
of Ligand B is 300nM, half the binding sites on Protein A
will be
filled.
E. When the concentration of Protein C is 10uM, half of
Ligand D will be bound
Protein A binds Ligand B with higher affinity than Protein C binds Ligand D
When the concentration of Ligand B is 300nM, half the binding sites on Protein A will be filled
Explain what characteristics of the protein and its ligand provides specificity of binding (i.e. what dictates the Kd of a protein-protein or protein-ligand interaction?)
The architecture of the bonds, as well as the number of bonds, formed between the atoms on the ligand and the atoms on the amino acids in the binding pocket of the protein
Which types of chemical bonds typically hold the protein and ligand together?
Non-covalent bonds like hydrophobic interactions, van der Waals,
hydrogen bonds and
electrostatic (ionic) bonds
A nuclear import peptide sequence can occur anywhere (inside or
outside) on a folded
protein, and most often is made up of a
string of charged amino acids such as -Lys-Lys-Lys-Arg-
Lys-.
FALSE; It is true that a nuclear import peptide typically has a
string of basic amino acids, but
they must be present on the
surface of a protein, not anywhere
GEF adds a phosphate to GDP to make GTP
FALSE; GEF triggers the release of GDP from a G-protein to allow GTP to bind in its place.
One of the ways that glycosylation is used in the ER is to mark
proteins for sorting to other
cellular compartments
TRUE; the sugar groups that are added during glycosylation are
continually modified as the
protein is made in the ER and passed
through the endocytic/secretion pathways
Post-translational modifications refer to
The formation of covalent bonds between protein subunits
The
addition of branched carbohydrates to a protein
A secreted protein, once translated, will follow which of the
following pathways through
the cell?
ER, vesicle, Golgi, vesicle, PM
hydrophobic effect
tendency of nonpolar molecules to clump together in water, while being excluded by water molecules
how to know if amino acid is polar
Side chains contain amide or hydroxyl groups
How to know if amino acid is non polar
Side chains are mostly hydrocarbon
how to know if amino acid is basic
Side chains contain amine functional groups
how to know amino acid is acidic
Side chains contain carboxyl groups
nonpolar amino acids
glycine, alanine, valine, cysteine, proline, leucine, isoleucine, methionine, trypyophan, phenylalnine
polar amino acids
serine, threonine, tyrosine, asparagine, glutamine
basic amino acids
lysine, arginine, histidine
acidic amino acids
aspartic acid, glutamic acid