Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
The flow of genetic information
front 1 How does RNA differ from DNA? | back 1 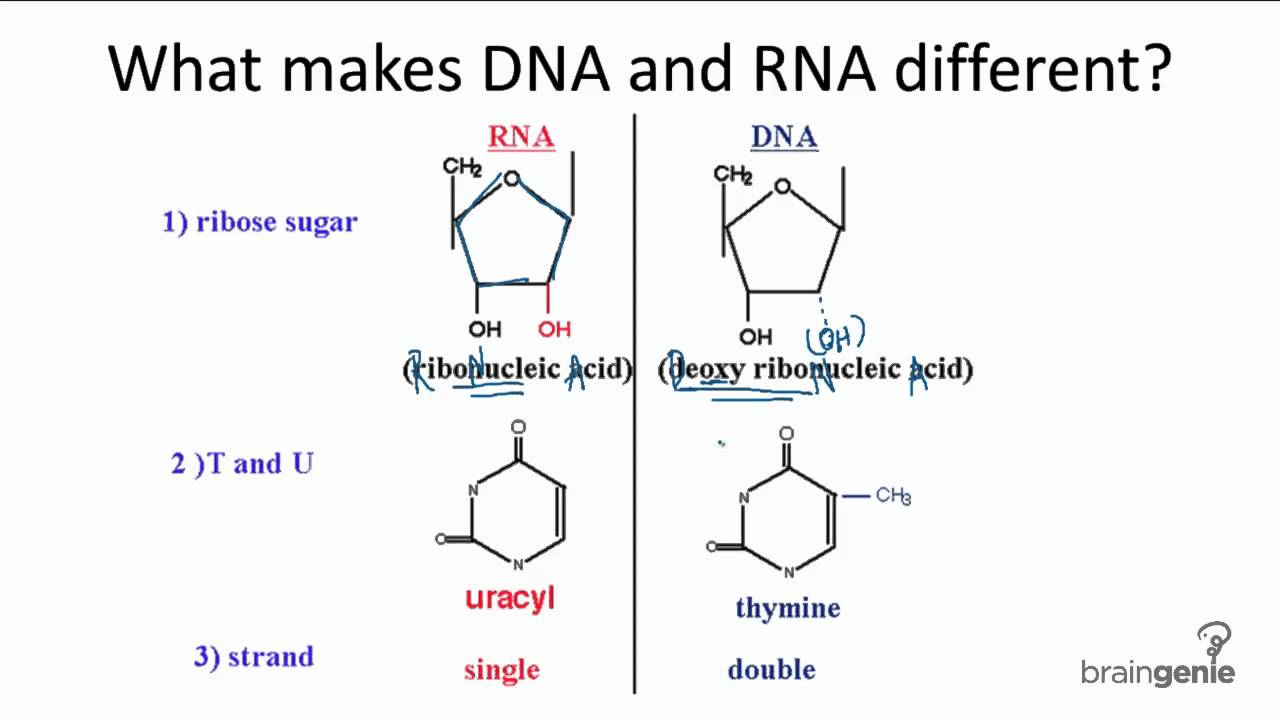
|
front 2  What are the three types or RNA? | back 2  |
front 3 What is transcription? | back 3 Copying of genetic information from the form of DNA to the form of RNA. Process that creates RNA from DNA using RNA polymerase. Think of it like this: you want to cook something, DNA is the cookbook that belongs to your friend(living in the nucleus), you can't take the book with you, so instead you copy the recipe down on a note card to take home(to the cytoplasm) (RNA.) |
front 4 What are the steps of transcription? | back 4 1. Initation 2. Elongation 3.Termination 4.Posttranscription modification |
front 5  What is a promoter? | back 5  Extra chunk of DNA sequence (located upstream 5' from the DNA region that contains the info to be transcribed into mRNA, and from the initiation start site. it doesn't code for anything, it is just a signal)
|
front 6 What is the template strand VS the coding strand | back 6
|
front 7 What is RNA Polymerase? | back 7 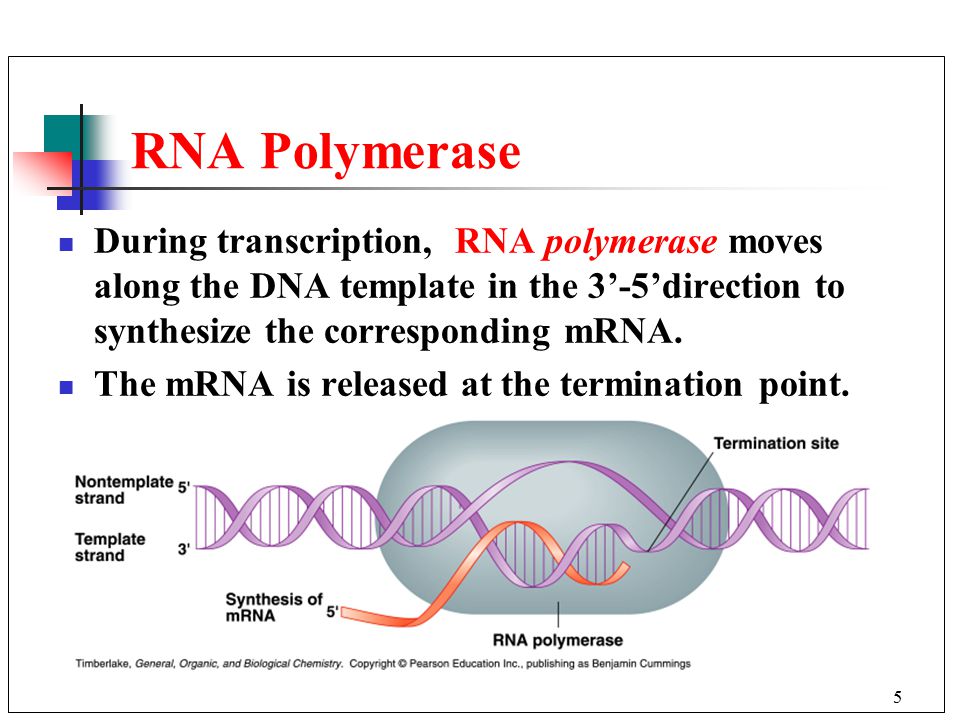 An enzyme that catalyzes the synthesis of an RNA strand from a DNA template. In eukaryotes, there are several classes of RNA polymerase. It assembles the individual nucleotides to create the RNA strand based on the DNA templet. It zips the DNA back up as it moves along the strand. It does not require a primer to imitate. |
front 8 In what direction does the RNA polymerase read the DNA template strand? In what direction does RNA polymerase synthesize and build | back 8 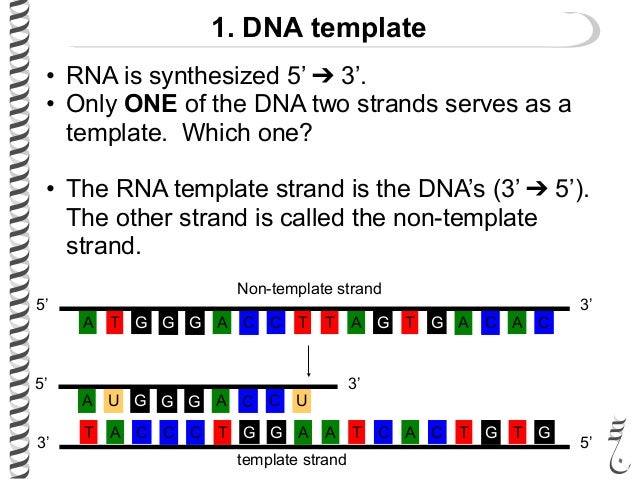
|
front 9 What are the RNA polymerase in eukaryotic cells | back 9  |
front 10 What is the job of RNA polymerase 1? | back 10 responsible for transcribing RNA that codes for genes that become structural components of the ribosome, a protein responsible for the translation of RNA into proteins. |
front 11 What is the job of RNA polymerase 2? | back 11 RNA polymerase II transcribes protein-encoding genes, or messenger RNAs, which are the RNAs that get translated into proteins |
front 12 What is the job of RNA polymerase 3? | back 12 transcribes a different structural region of the ribosome, transfer RNAs, which are also involved the translation process, as well as non-protein encoding RNAs. |
front 13 What is RNA polymerase in bacteria | back 13  Called holoenzyme and contains many subunits
|
front 14 What part of the RNA polymerase molecule is responsible for recognizing the promoter region of DNA in bacteria? | back 14 The sigma subunit |
front 15 Can eukaryotic polymerase recognize a promoter region easily? | back 15 NO, eukaryotic polymerases are unable to recognize promoter regions. They have no direct parallel to the sigma subunit of their prokaryotic counterpart. Instead, eukaryotic polymerases depend on other proteins that bind to the promoter regions and then recruit the RNA polymerases to the correct spots. |
front 16 What do eukaryotic polymerase use to recognize a promoter region? | back 16 the TATA box, the CAAT box, or the GC box, which all act as binding sites for specific transcription factors that will then recruit the correct polymerase
|
front 17 What are transcription factors? | back 17 Transcription factors are proteins involved in the process of converting, or transcribing, DNA into RNA. Transcription factors include a wide number of proteins, excluding RNA polymerase, that initiate and regulate the transcription of genes. |
front 18 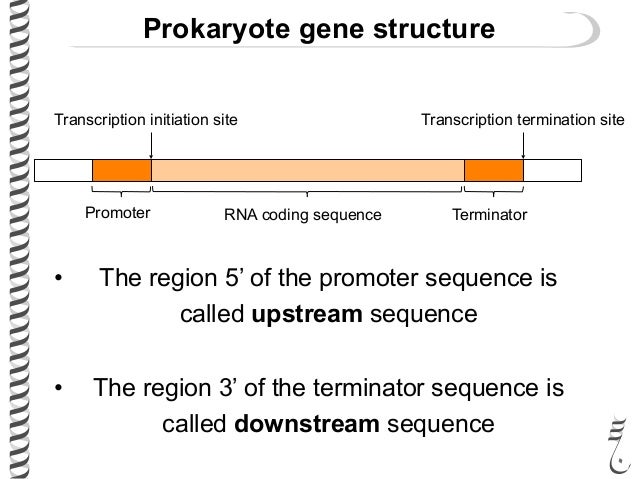 Terms upstream and downstream are commonly used when discussing the location of specific loci | back 18  |
front 19  Examples of census sequences | back 19 TATA box is a DNA sequence located on the nontemplate strand, upstream from transcription start site. that indicates where a genetic sequence can be read and decoded. It is a type of promoter sequence, which specifies to other molecules where transcription begins. TATATA rich because easier to break apart because of 2 hydrogen bonds. CCAAT box (also sometimes abbreviated a CAAT box or CAT box) is a distinct pattern of nucleotides with GGCCAATCT consensus sequence that occur upstream by 60-100 bases to the initial transcription site. Both are Cis acting elements. The Pribnow box (also known as the Pribnow-Schaller box) is the sequence TATAAT of six nucleotides (thymine-adenine-thymine-etc.) that is an essential part of a promoter site on DNA for transcription to occur in bacteria. located -10 When we give consensus sequences, we give the sequence of the coding strand. For example, the Pribnow box (-10) sequence is: 5’ TATAAT 3’ |
front 20
| back 20 positioned at -35 and -10 with respect to the transcription initiation site. |
front 21 What would happen if a mutation occurred in the consensus sequences? | back 21 transcription levels would decrease |
front 22 Cis acting vs trans acting elements | back 22 Cis-regulatory elements are often binding sites for one or more trans-actingfactors.
|
front 23 Do all promoters have the same CIs elements | back 23 NO |
front 24 What takes place in the first step on transcription: initation | back 24 
|
front 25 Is a primer necessary for RNA polymerase to start synthesizing in the second step of transcription known as elongation? | back 25 NO, not necessary, the RNA polymerase just keeps synthesizing and adding to that 3 prime end of mRNA during elongation |
front 26 For a given gene, one strand serves as the template, but... | back 26 either strand can be the template depending on which direction(left or right) it says the DNA is being read or the RNA is being synthesized. Different strands of the double helix DNA are used as the template for different genes The location of the promoter will dictate the site of RNA Polymerase binding and direction of transcription |
front 27 RNA polymerase stops transcribing when what happens? | back 27 When it reaches the termination sequence. It is different in prokaryotic bacteria and eukaryotic |
front 28 How does termination happen in prokaryotes? | back 28 RNA synthesis will continue along the DNA template strand until the polymerase encounters a signal that tells it to stop, or terminate, transcription. In prokaryotes, this signal can take two forms, rho-independent and rho-dependent. |
front 29 What is the rho-independent terminator in bacteria? (intrinsic) | back 29  RNA can fold back and base pair with itself forming a hairpin loop. the string of adenines in the DNA sequence are transcribed into uracils in the RNA sequence. Because the uracil bases will only pair weakly with the adenines, the RNA chain can easily be released from the DNA template, terminating transcription. |
front 30 In the hairpin loop, what is the purpose of the strong GC bonds before the loop? | back 30
|
front 31 What is rho-dependent termination in bacteria? | back 31 rho-dependent terminator received its name because it is dependent on a specific protein called a rho factor. The rho factor is thought to bind to the end of the RNA chain and slide along the strand towards the open complex bubble. When the factor catches the polymerase, it causes the termination of transcription. |
front 32 How is the chromatin different in prokaryotes vs eukaryotes in regards to transcription? What about Cis acting elements and transacting factors? | back 32
|
front 33 Where are enhancers and silencers located in eukaryotic? | back 33 RNA rely on transcription factors, in addition to promoters, other control units enhancers and silencers that can be located in the 5' regulatory region upstream from initation point or 3' downstream region beyond coding sequence |
front 34 Why needs to happen before RNA transcripts can be transported to the cytoplasm | back 34 They require significant alteration to become mature mRNA before they're transported to the cytoplasm, via post transcriptional modification |
front 35 What takes place in postranscriptional modification? | back 35
|
front 36 If the 5’cap or poly A tail were missing from a eukaryotic mRNA, what would be the consequence | back 36  More prone to degradation/ lack of transport to cytoplasm |
front 37 What is mRNA splicing? | back 37
|
front 38 How does mRNA splicing occur? | back 38  via spicisomes which are built up of small proteins+snRNA which would be found in the nucleus of a cell |
front 39 What are the products of eukaryotic transcription compared to prokaryotic? | back 39 Eukaryotic transcription yields pre-mRNA or primary transcript RNA. Prokaryotic transcription yields mature RNA (mRNA) |
front 40 What do the splicisomes do? | back 40  recognizes specific sequences in the DNA that will determine the sites of splicing |
front 41 What is the name of protein coding segments of primary transcript RNA? Of non- protein coding segments? Which are conserved during the splicing reaction? | back 41 Exons are segments of primary transcript RNA that are conserved during the splicing reaction and code for proteins during translation. Introns are sequences that are not conserved and are spliced out during the splicing reaciton. **UTR does NOT get spliced out!!!! |
front 42 How many different genetic combinations can be made from the 3 nuleodite codons and how many amino acids do they code for? | back 42
|
front 43
| back 43 
|
front 44 What is translation? | back 44 process that converts an mRNA sequence into a string of amino acids that form a protein |
front 45 What are the 3 main steps of translation? | back 45
|
front 46 What is the anticodon? | back 46  The sequence of three nucleotides located on the anticodon arm of the tRNA cloverleaf structure, the tRNA brings this anticodon with it. The anticodon bonds in antiparallel fashion with a codon of mRNA at the acceptor site of a ribosome during translation. This is like a lock and key, its how the tRNA knows its at the right spot, when the key fits! |
front 47 Before translation can occur, what does tRNA need to be bound with? | back 47 An amino acid in two-step process is called "charging". |
front 48 What is Adenylylation? | back 48 The first step in tRNA charging. Involves the "activation" of an amino acid so that the acid can be bound to a tRNA molecule. The process of activation involves the transfer of an AMP group from ATP to the amino acid. |
front 49 Aminoacyl-tRNA synthase | back 49
|
front 50 Aminoacyl tRNA | back 50 A tRNA molecule that has been charged. It is loaded with an amino acid and is ready to participate in translation at the ribosome, where it binds to the acceptor site. Also known as charged tRNA |
front 51 What is charging? | back 51 The two-step process in which an amino acid is "loaded" onto a tRNA. The first step is adenylylation; the second is the binding of tRNA and amino acid into an aminoacyl tRNA. |
front 52 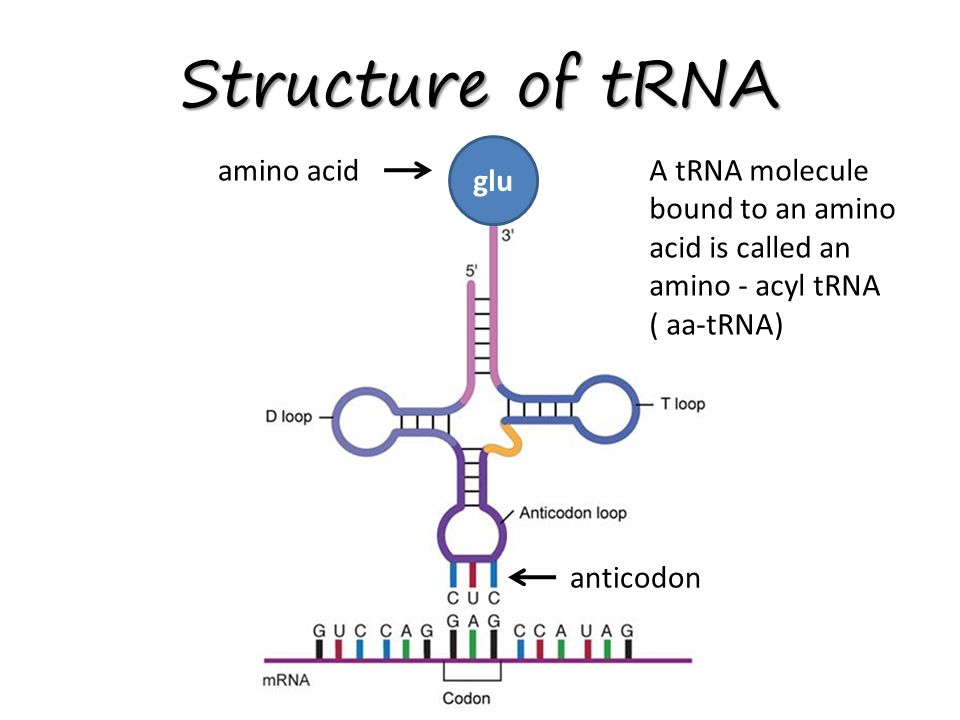 What is located at the 3' end of the tRNA? | back 52  At the 3' end of the tRNA molecule, opposite the anticodon, extends a three nucleotide acceptor site (that includes a free -OH group) known as the amino acid attachment site. A specific tRNA binds to a specific amino acid through its acceptor stem. |
front 53 What do the ribosomes work as? | back 53  “as a nonspecific workbench for the translation process.” |
front 54 What is rRNA's job in translation? | back 54 to assist in the binding processes between tRNA anticodon and mRNA codon |
front 55 So what happens in initiation in translation? | back 55
|
front 56 When does codon recognition happen? | back 56 Codon recognition happens when tRNA pairs with the mRNA inside the ribosome. example: AUG on the mRNA is the start codon, the anticodon on the tRNA that matches it would be UAC, and it codes for methionine |
front 57 What does the chemical structure of an amino acid look like? | back 57  chemical structure of an amino acid, we see that one end contains a terminal nitrogen group while the other contains a carboxyl group. |
front 58 Whats happening in elongation? | back 58 
|
front 59 What is each amino acid being added called? | back 59  each amino acid added can be called a peptide; a building block of the larger polypeptide chain. Proteins are polypeptides. |
front 60 What type of bond does addition occur through? | back 60 addition occurs through the formation of a peptide bond, the nitrogen-carbon bond that forms between amino acid subunits to form a polypeptide chain |
front 61 Protein chain growth occurs in what direction? | back 61 N-terminus to carboxy-terminus |
front 62 In which direction does the ribosome move along the mRNA molecule | back 62
|
front 63 When does translation end and termination occur? | back 63 Translation ends when one of three stop codons, UAA, UAG, or UGA, enters the A site of the ribosome |
front 64 Does a stop codon code for anything? UGA,UAG,UAA? | back 64 The three stop codons are UAA, UAG, and UGA. These codons signal translation termination because there is no tRNA molecule that recognizes these codons. Instead, they are recognized by termination factors that stop translation. |