Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
Exam 3: Ch. 17: From Gene to Protein
front 1 What is the start codon? | back 1 AUG Nearly every mRNA gene that codes for a protein begins with the start codon, AUG, and thus begins with a methionine. |
front 2 What are the stop codons? | back 2 UAG, UGA, UAA Nearly every protein-coding sequence ends with one of the three stop codons (UAA, UAG, and UGA), which do not code for amino acids but signal the end of translation. |
front 3 During translation, nucleotide base triplets (codons) in mRNA are read in sequence in the 5’ → 3’ direction along the mRNA. Amino acids are specified by the string of codons. What amino acid sequence does the following mRNA nucleotide sequence specify? 5′−AUGGCAAGAAAA−3′ Express the sequence of amino acids using the three-letter abbreviations, separated by hyphens (e.g., Met-Ser-Thr-Lys-Gly). | back 3 Met-Ala-Arg-Lys An amino acid sequence is determined by strings of three-letter codons on the mRNA, each of which codes for a specific amino acid or a stop signal. The mRNA is translated in a 5’ → 3’ direction. |
front 4 Which amino acid does the codon GCA code for? | back 4 Ala |
front 5 To identify the amino acids specified by the mRNA sequence, you first need to subdivide the sequence into codons of three nucleotides each. This can be done by placing a space between each codon. Which of the following is the correct division of the codons for the sequence given? Look for the correct placement of spaces. A. 5′−A UGG CAA GAA AA−3′ B. 5′−AU GGC AAG AAA A−3′ C. 5′−AUG GCA AGA AAA−3′ D. 5′−AUGG CAA GAA AA−3′ E. 5′−AU GGCA AGAA AA−3′ | back 5 C. 5′−AUG GCA AGA AAA−3′ |
front 6 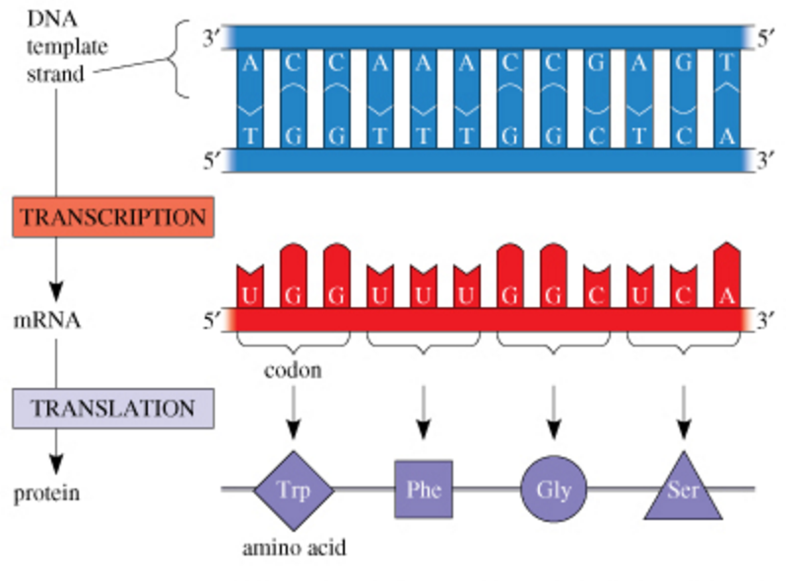 The role of DNA in determining amino acid sequences Before a molecule of mRNA can be translated into a protein on the ribosome, the mRNA must first be transcribed from a sequence of DNA. What amino acid sequence does the following DNA nucleotide sequence specify? 3′−TACAGAACGGTA−5′ Express the sequence of amino acids using the three-letter abbreviations, separated by hyphens (e.g., Met-Ser-His-Lys-Gly). | back 6 Met-Ser-Cys-His Before mRNA can be translated into an amino acid sequence, the mRNA must first be synthesized from DNA through transcription. Base pairing in mRNA synthesis follows slightly different rules than in DNA synthesis: uracil (U) replaces thymine (T) in pairing with adenine (A). The codons specified by the mRNA are then translated into a string of amino acids. |
front 7 Steps to convert a DNA sequence into an amino acid sequence.
| back 7 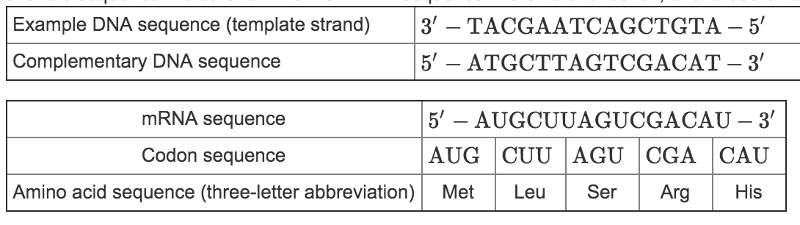 Sample Problem: This chart shows how to decode an example DNA sequence. Remember to first determine the mRNA sequence that is complementary to the DNA template strand’s sequence. Be sure to write the mRNA sequence in a 5’ to 3’ direction, and to use U to pair with A. |
front 8 The flow of information in a cell proceeds in what sequence? A. from DNA to RNA to protein B. from RNA to DNA to protein C. from RNA to protein to DNA D. from DNA to protein to RNA E. from protein to RNA to DNA | back 8 A. from DNA to RNA to protein This is known as the central dogma of biology. |
front 9 A codon consists of _____ bases and specifies which _____ will be inserted into the polypeptide chain. A. four ... fatty acid B. two ... nucleotide C. three ... nucleotide D. three ... amino acid E. four ... amino acid | back 9 D. three ... amino acid Three nucleotide bases make up a codon and specify which amino acid comes next in the sequence. |
front 10 A particular triplet of bases in the template strand of DNA is 5' AGT 3'. The corresponding codon for the mRNA transcribed is _____. A. 5' TCA 3' B. 3' UGA 5' C. 3' ACU 5' D. 3' UCA 5' | back 10 D. 3' UCA 5' |
front 11 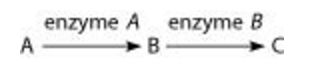 The figure above shows a simple metabolic pathway. According to Beadle and Tatum's hypothesis, how many genes are necessary for this pathway? A. 1 B. 2 C. 3 D. It cannot be determined from the pathway. | back 11 B. 2 |
front 12 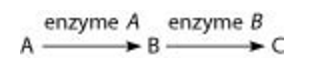 Refer to the metabolic pathway illustrated above. If A, B, and C are all required for growth, a strain that is mutant for the gene-encoding enzyme A would be able to grow on medium supplemented with _____. A. nutrient A only B. nutrient B only C. nutrient C only D. nutrients A and C | back 12 B. nutrient B only |
front 13 In the process of transcription, _____. A. DNA is replicated B. mRNA attaches to ribosomes C. RNA is synthesized D. proteins are synthesized | back 13 C. RNA is synthesized |
front 14 Which of the following specifies a single amino acid in a polypeptide chain? A. the three-base sequence of mRNA B. the complementarity of DNA and RNA C. the aminoacyl-tRNA synthetase D. the base sequence of the tRNA | back 14 A. the three-base sequence of mRNA |
front 15  In the diagram, the gray unit represents _____. A. RNA B. DNA C. transcription factors D. RNA polymerase E. The promoter | back 15 D. RNA polymerase RNA polymerase untwists a portion of the DNA double helix. |
front 16  In the diagram, the green unit represents _____. A. RNA B. DNA C. transcription factors D. RNA polymerase E. The promoter | back 16 E. The promoter The promoter is the region of DNA at which the process of transcription begins. |
front 17  In the diagram below, the two blue strands represent _____. A. RNA B. DNA C. transcription factors D. RNA polymerase E. The promoter | back 17 B. DNA RNA is not a double helix. DNA is a double helix. |
front 18  Which of these correctly illustrates the pairing of DNA and RNA nucleotides? | back 18 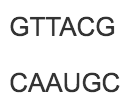 In RNA, uracil takes the place of thymine. |
front 19 The direction of synthesis of an RNA transcript is _____. A. 1' —> 5' B. 5' —> 3' C. 1' —> 3' D. 3' —> 5' E. 2' —> 4' | back 19 B. 5' —> 3' Nucleotides are added to the 3' end of RNA. |
front 20 Where does RNA polymerase begin transcribing a gene into mRNA? A. It starts at one end of the chromosome. B. The ribosome directs it to the correct portion of the DNA molecule. C. Transfer RNA acts to translate the message to RNA polymerase. D. It starts after a certain nucleotide sequence called a promoter. E. It looks for the AUG start codon. | back 20 D. It starts after a certain nucleotide sequence called a promoter. Remember that RNA polymerase is an enzyme. In both eukaryotes and prokaryotes, RNA polymerase binds to the gene's promoter and begins transcription at a nucleotide known as the start point, although in eukaryotes the binding of RNA polymerase to the promoter requires transcription factors. |
front 21 Transcription in eukaryotes requires which of the following in addition to RNA polymerase? A. aminoacyl-tRNA synthetase B. several transcription factors C. start and stop codons D. ribosomes and tRNA | back 21 B. several transcription factors |
front 22 During RNA processing a(n) _____ is added to the 5' end of the RNA. A. 3' untranslated region B. a long string of adenine nucleotides C. 5' untranslated region D. coding segment E. modified guanine nucleotide | back 22 E. modified guanine nucleotide The 5' cap consists of a modified guanine nucleotide. |
front 23 During RNA processing a(n) _____ is added to the 3' end of the RNA. A. 3' untranslated region B. a long string of adenine nucleotides C. 5' untranslated region D. coding segment E. modified guanine nucleotide | back 23 B. a long string of adenine nucleotides A poly-A tail is added to the 3' end of the RNA. |
front 24 Spliceosomes are composed of _____. A. snRNPs and other proteins B. polymerases and ligases C. introns and exons D. the RNA transcript and protein E. snRNPs and snurps | back 24 A. snRNPs and other proteins These are the component of spliceosomes. |
front 25 The RNA segments joined to one another by spliceosomes are _____. A. caps B. exons C. snRNPs D. tails E. introns | back 25 B. exons Exons are expressed regions. |
front 26 Translation occurs in the _____. A. cytoplasm B. lysosome C. nucleus D. Golgi apparatus E. nucleoplasm | back 26 A. cytoplasm Ribosomes, the sites of translation, are found in the cytoplasm. |
front 27 After an RNA molecule is transcribed from a eukaryotic gene, what are removed and what are spliced together to produce an mRNA molecule with a continuous coding sequence? A. silencers ... enhancers B. promoters ... operators C. exons ... introns D. operators ... promoters E. introns ... exons | back 27 E. introns ... exons This RNA processing does not occur in bacterial cells. Introns, intervening sequences, are removed and the exons, expressed sequences, are spliced together. |
front 28 Alternative RNA splicing _____. A. increases the rate of transcription B. can allow the production of proteins of different sizes and functions from a single gene C. can allow the production of similar proteins from different RNAs D. is a mechanism for increasing the rate of translation | back 28 B. can allow the production of proteins of different sizes and functions from a single gene |
front 29 Use this model of a eukaryotic transcript to answer the following
question. E = exon and I = intron A. 5' UTR I1 I2 I3 UTR 3' B. 5' E1 I1 E2 I2 E3 I3 E4 3' C. 5' UTR E1 E2 E3 E4 UTR 3' D. 5' E1 E2 E3 E4 3' | back 29 C. 5' UTR E1 E2 E3 E4 UTR 3' |
front 30 Locations of the processes involved in protein synthesis In eukaryotic cells, the processes of protein synthesis occur in different cellular locations. Drag the labels to the appropriate targets to identify where in the cell each process associated with protein synthesis takes place. | back 30 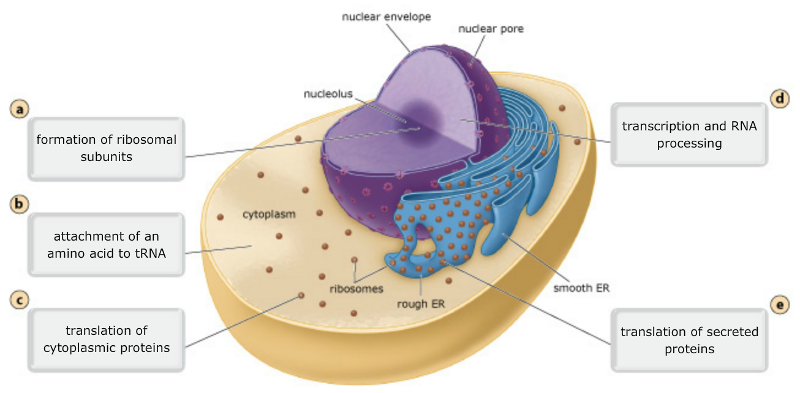 |
front 31 What occurs during some key processes of protein synthesis? Match these key processes involved in protein synthesis to descriptions of what occurs at each step. | back 31 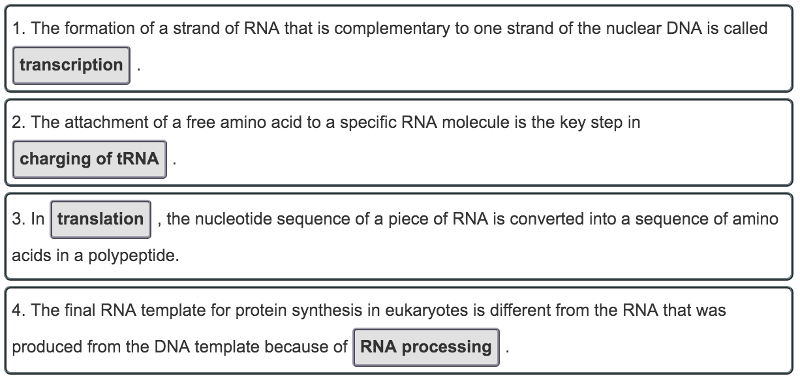 |
front 32 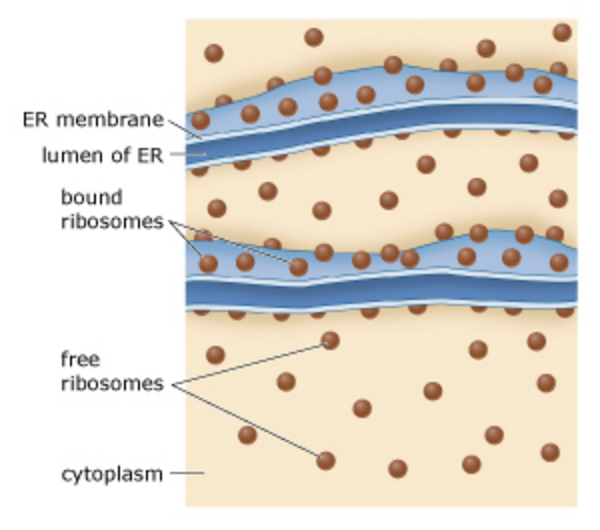 Where are cytoplasmic and secreted proteins made? Both cytoplasmic and secreted proteins can only be synthesized in the presence of a ribosome. This diagram shows the two kinds of ribosomes:
| back 32 NO QUESTION |
front 33 Which statement correctly describes where cytoplasmic and secreted proteins are synthesized? A. Both cytoplasmic and secreted proteins are synthesized on free ribosomes. B. Cytoplasmic proteins are synthesized on free ribosomes, whereas secreted proteins are synthesized on ribosomes bound to the rough ER. C. Cytoplasmic proteins are synthesized on ribosomes bound to the rough ER, whereas secreted proteins are synthesized on free ribosomes. D. Both cytoplasmic and secreted proteins are synthesized on ribosomes bound to the rough ER. | back 33 B. Cytoplasmic proteins are synthesized on free ribosomes, whereas secreted proteins are synthesized on ribosomes bound to the rough ER. |
front 34 Roles of RNA in protein synthesis in eukaryotes RNA plays important roles in many cellular processes, particularly those associated with protein synthesis: transcription, RNA processing, and translation. Drag the labels to the appropriate bins to identify the step in protein synthesis where each type of RNA first plays a role. If an RNA does not play a role in protein synthesis, drag it to the “not used in protein synthesis” bin. | back 34 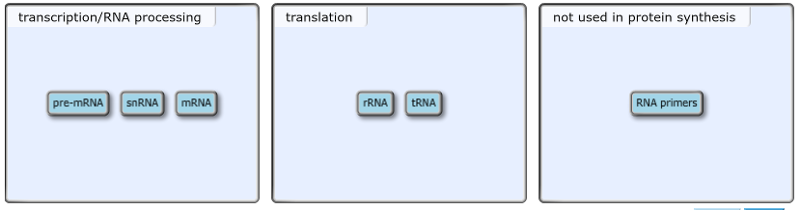 In eukaryotes, pre-mRNA is produced by the direct transcription of the DNA sequence of a gene into a sequence of RNA nucleotides. Before this RNA transcript can be used as a template for protein synthesis, it is processed by modification of both the 5' and 3' ends. In addition, introns are removed from the pre-mRNA by a splicing process that is catalyzed by snRNAs (small nuclear RNAs) complexed with proteins. The product of RNA processing, mRNA (messenger RNA), exits the nucleus. Outside the nucleus, the mRNA serves as a template for protein synthesis on the ribosomes, which consist of catalytic rRNA (ribosomal RNA) molecules bound to ribosomal proteins. During translation, tRNA (transfer RNA) molecules match a sequence of three nucleotides in the mRNA to a specific amino acid, which is added to the growing polypeptide chain. RNA primers are not used in protein synthesis. RNA primers are only needed to initiate a new strand of DNA during DNA replication. |
front 35 The role of RNA primers DNA synthesis (replication) and RNA synthesis differ in their needs for primer molecules.
| back 35 NO QUESTION |
front 36 How do tRNA and rRNA function in protein synthesis? Both tRNA (transfer RNA) and rRNA (ribosomal RNA) play essential roles in protein synthesis. Which two statements correctly describe the roles of tRNA and rRNA in protein synthesis? A. rRNA is the major structural component of ribosomes and is involved in binding both mRNA and tRNAs. B. tRNAs implement the genetic code, translating information from a sequence of nucleotides to the sequence of amino acids that make up a protein. C. tRNA transfers a nucleotide sequence from the DNA in the nucleus to the site of protein synthesis in the cytoplasm. D. rRNA has many variations, each of which binds a specific amino acid. | back 36 A. & B. |
front 37 What is the role of mRNA in protein synthesis? mRNA (messenger RNA) plays a key role in protein synthesis as the intermediate between the information encoded by a sequence of bases in DNA (a gene) and the sequence of amino acids that make up the protein product. Which three statements correctly describe the role that mRNA plays in protein synthesis in eukaryotes? A. mRNA is produced only after the steps of RNA processing. B. mRNA is the template for protein synthesis in translation. C. mRNA links together amino acids, forming a polypeptide chain. D. mRNA carries genetic information from the nucleus to the cytoplasm. E. mRNA is the immediate product of transcription. | back 37 A., B., & D. |
front 38 snRNAs and RNA processing One stage of RNA processing in eukaryotes involves the removal of introns--non-coding regions interspersed within the coding regions of the pre-mRNA. In this RNA splicing process, the machinery that catalyzes the removal of introns (called the spliceosome) is composed of proteins and snRNAs (small nuclear RNAs). The snRNAs (and associated proteins) have two functions in the splicing process:
| back 38 NO QUESTION |
front 39  Martian Part 1 Life as we know it depends on the genetic code: a set of codons, each made up of three bases in a DNA sequence and corresponding mRNA sequence, that specifies which of the 20 amino acids will be added to the protein during translation. Imagine that a prokaryote-like organism has been discovered in the polar ice on Mars. Interestingly, these Martian organisms use the same DNA → RNA → protein system as life on Earth, except that
Based on this information, what is the minimum size of a codon for these hypothetical Martian life-forms? A. 2 bases B. 3 bases C. 4 bases D. 5 bases E. 6 bases F. The answer cannot be determined from the information provided. | back 39 D. 5 bases In the most general case of x bases and y bases per codon, the total number of possible codons is equal to xy . In the case of the hypothetical Martian life-forms, is the minimum codon length needed to specify 17 amino acids is 5 (25 = 32), with some redundancy (meaning that more than one codon could code for the same amino acid). For life on Earth, x = 4 and y = 3; thus the number of codons is 43, or 64. Because there are only 20 amino acids, there is a lot of redundancy in the code (there are several codons for each amino acid). |
front 40 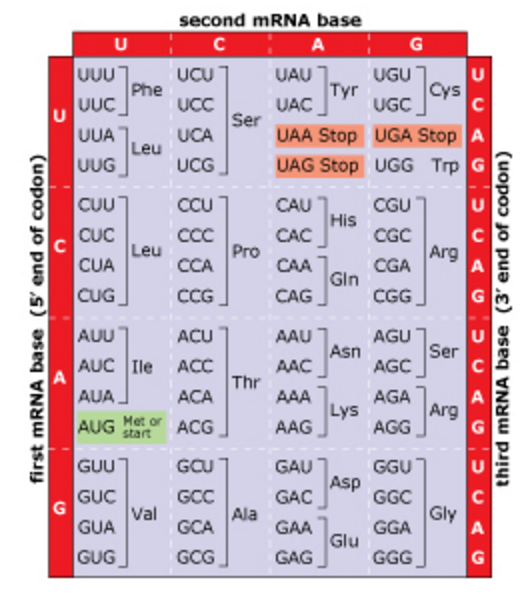 Martian Part 2 A simple mathematical equation can correctly express the maximum number of codons that can be constructed from x different bases, with a codon length ofy bases. Recall that for life on Earth,
Which of the following equations can be used to calculate the maximum number of codons (N) that can be constructed from x different bases when there are y bases per codon? A. N = x y B. N = x y C. N = 4x(y+1) D. N = 16(3x/y) | back 40 A. N = x y |
front 41 Martian Part 3 It is possible to solve this problem by simply listing all the possible combinations of bases for a given codon length. In the hypothetical Martian example there are only 2 different bases (A and T). For a codon length of 1 base, there would only be 2 possible codons: A or T For a codon length of 2 bases, there would be 4 possible codons: AA; TA; AT; or TT How many different Martian codons are possible if the codon length is 4 bases? A. 12 B. 16 C. 32 D. 64 | back 41 B. 16 |
front 42 These three events happen every time mRNA is translated into a protein. Put these events in order. Rank from first to last. To rank items as equivalent, overlap them. Translation Initiation Termination | back 42 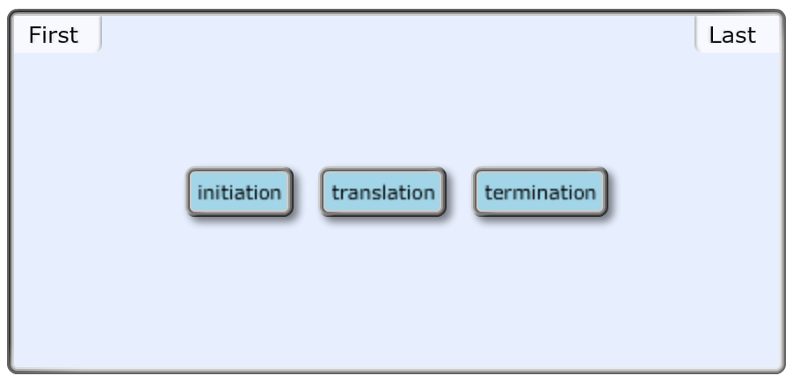 |
front 43 What is the the function of tRNA? A. It binds the mRNA before the ribosome and guides the ribosome to the correct binding place. B. It recognizes and binds a specific codon on mRNA during translation. C. It contains the gene coding for a protein. | back 43 B. It recognizes and binds a specific codon on mRNA during translation. |
front 44 What is the function of aminoacyl-tRNA synthetase? A. It synthesizes amino acids. B. It attaches amino acids to a specific tRNA. C. It synthesizes tRNA. | back 44 B. It attaches amino acids to a specific tRNA. |
front 45 Complete the following vocabulary exercise related to the process of translation of mRNA to protein by the ribosome. Match the words in the left-hand column with the appropriate blank in the sentences in the right-hand column. | back 45 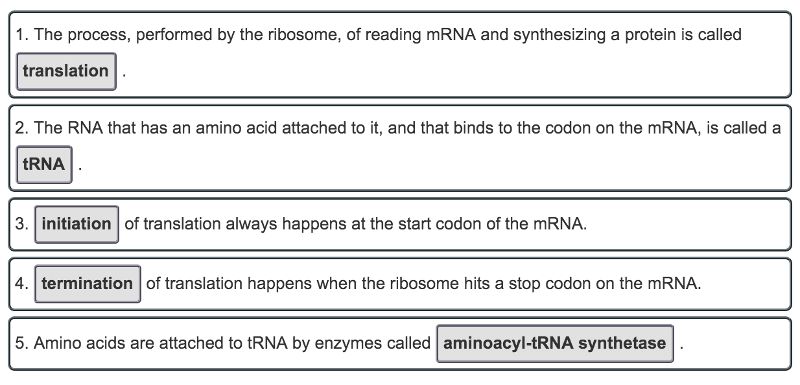 The process of translation, or protein synthesis, is a crucial part of the maintenance of living organisms. Proteins are constantly in use and will break down eventually, so new ones must always be available. If protein synthesis breaks down or stops, then the organism dies. |
front 46 A particular triplet of bases in the coding sequence of DNA is AAA. The anticodon on the tRNA that binds the mRNA codon is _____. A. UUA B. UUU C. AAA D. TTT | back 46 B. UUU |
front 47 Accuracy in the translation of mRNA into the primary structure of a polypeptide depends on specificity in the _____. A. binding of the anticodon to small subunit of the ribosome B. binding of ribosomes to mRNA C. attachment of amino acids to rRNAs D. binding of the anticodon to the codon and the attachment of amino acids to tRNAs | back 47 D. binding of the anticodon to the codon and the attachment of amino acids to tRNAs |
front 48 The release factor (RF) _____. A. supplies a source of energy for termination of translation B. releases the ribosome from the ER to allow polypeptides into the cytosol C. binds to the stop codon in the A site in place of a tRNA D. releases the amino acid from its tRNA to allow the amino acid to form a peptide bond | back 48 C. binds to the stop codon in the A site in place of a tRNA |
front 49 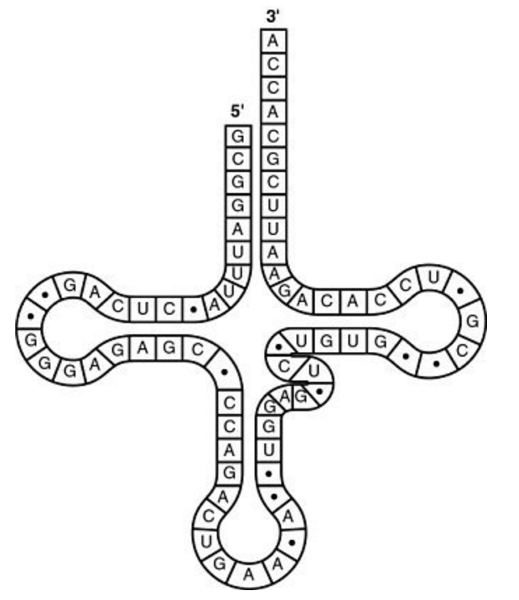 What type of bonding is responsible for maintaining the shape of the tRNA molecule shown in the figure above? A. hydrogen bonding between base pairs B. ionic bonding between phosphates C. peptide bonding between amino acids D. van der Waals interactions between hydrogen atoms | back 49 A. hydrogen bonding between base pairs |
front 50  The tRNA shown in the figure above has its 3' end projecting beyond its 5' end. What will occur at this 3' end? A. The amino acid binds covalently. B. The excess nucleotides (ACCA) will be cleaved off at the ribosome. C. The 5' cap of the mRNA will become covalently bound. D. The small and large subunits of the ribosome will attach to it. | back 50 A. The amino acid binds covalently. |
front 51 During elongation, which site in the ribosome represents the location where a codon is being read? A. E site B. A site C. P site D. the small ribosomal subunit | back 51 B. A site |
front 52 How does termination of translation take place? A. The end of the mRNA molecule is reached. B. A stop codon is reached. C. The 5' cap is reached. D. The poly-A tail is reached. | back 52 B. A stop codon is reached. |
front 53 Which of the following types of mutation, resulting in an error in the mRNA just after the AUG start of translation, is likely to have the most serious effect on the polypeptide product? A. a substitution of the third nucleotide in an ACC codon B. a deletion of two nucleotides C. a substitution of the first nucleotide of a GGG codon D. a deletion of a codon | back 53 B. a deletion of two nucleotides |
front 54 What does a mutagen cause? Consider the meaning of the prefix "muta-." A. decreased enzyme activity throughout the cell B. a change in the sequence of DNA C. decreased permeability of the nuclear envelope D. problems with mitosis E. a reduction in the number of tRNA molecules available for protein synthesis | back 54 B. a change in the sequence of DNA Mutations are changes in the genetic material of the cell. |
front 55 A nonsense mutation in a gene _____. A. changes an amino acid in the encoded protein B. has no effect on the amino acid sequence of the encoded protein C. introduces a premature stop codon into the mRNA D. alters the reading frame of the mRNA | back 55 C. introduces a premature stop codon into the mRNA |
front 56 The most commonly occurring mutation in people with cystic fibrosis is a deletion of a single codon. This results in _____. A. a polypeptide missing an amino acid B. a frameshift mutation C. a base-pair substitution D. a nonsense mutation | back 56 A. a polypeptide missing an amino acid |
front 57 How might a single base substitution in the sequence of a gene affect the amino acid sequence of a protein encoded by the gene, and why? A. Only a single amino acid could change, because the reading frame is unaffected. B. The amino acid sequence would be substantially altered, because the reading frame would change with a single base substitution. C. All amino acids following the substitution would be affected, because the reading frame would be shifted. D. It is not possible for a single base substitution to affect protein structure, because each codon is three bases long. | back 57 A. Only a single amino acid could change, because the reading frame is unaffected. |