Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
Genetics exam 2
front 1 A document to record ancestry, used by genealogists in study of human family lines, and in selective breeding of other animals | back 1 Pedigree |
front 2 Why is human genetics hard to follow? | back 2 -a small number of offspring -experimental crosses not ethical -long generation time (22-23 yrs) |
front 3 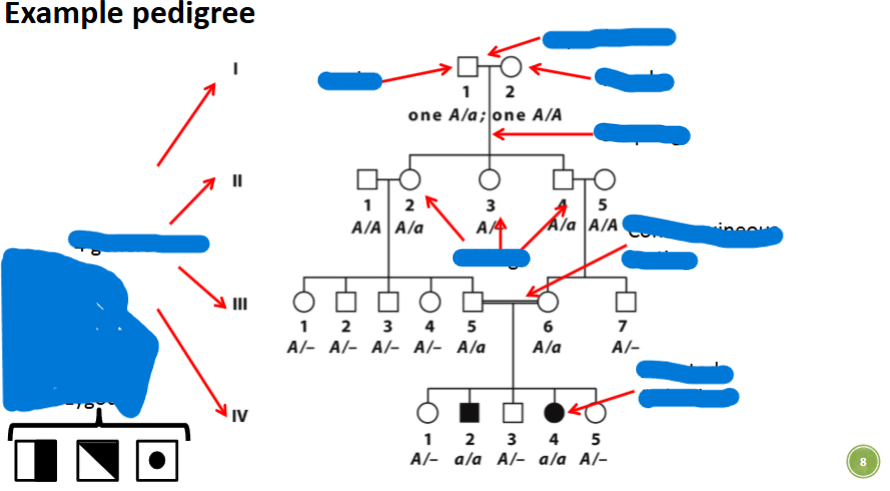 | back 3 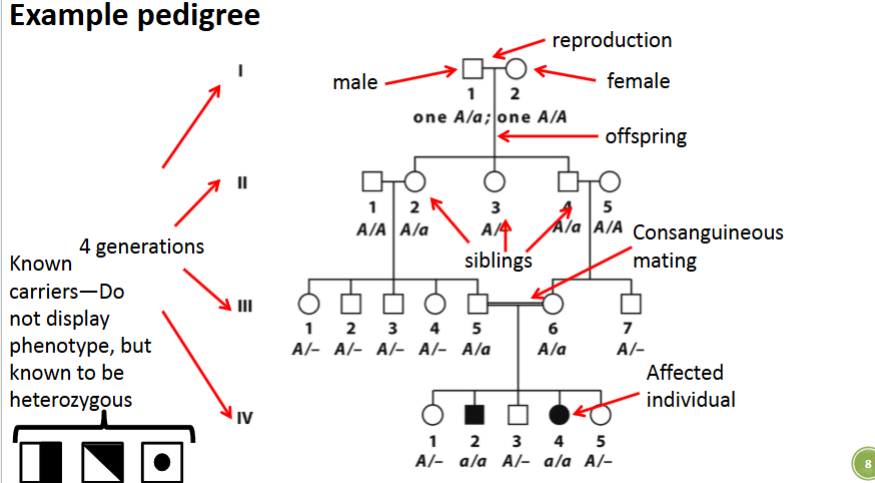 |
front 4 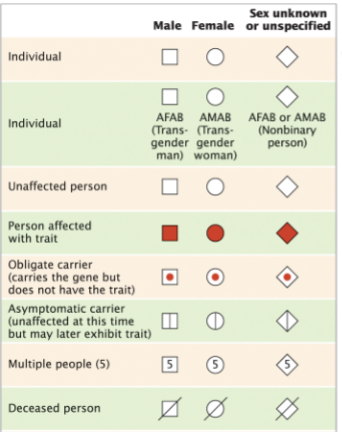 | back 4 .. |
front 5 What are the 4 basic inheritance patterns? | back 5 -Autosomal recessive -Autosomal dominant - Sex-linked recessive -Sex-linked dominant |
front 6 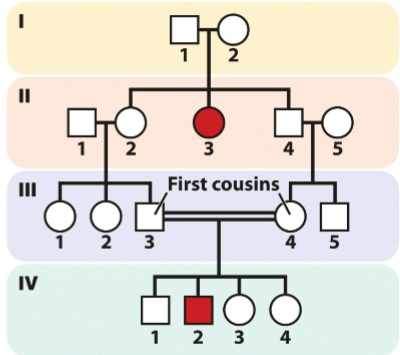 -2 unaffected parents can have affected offspring -both male and female offspring affected at similar freq. | back 6 Autosomal recessive |
front 7 What are examples of autosomal recessive disorders? | back 7 -cystic fibrosis (severe damage to lungs)(causes thickened fluids) -Sickle cell anemia (affects shape of blood cell) -Tay-Sachs (toxic build up in brain and spinal cord) |
front 8 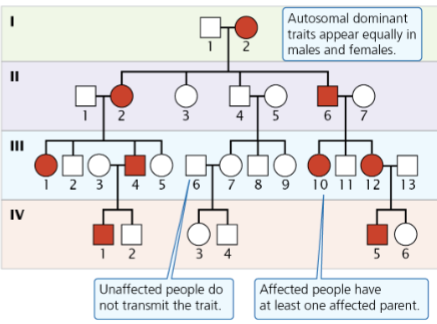 -Affected individuals have at least 1 affected parent -Affected parent of either sex can pass on to offspring of either sex -appears in male and female - not sex linked | back 8 Autosomal dominant |
front 9 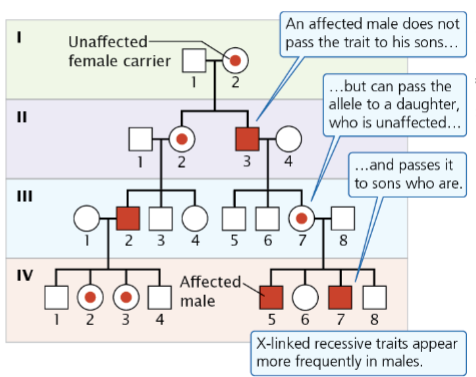 -unaffected parents can have affected offspring -Males affected more frequently than females -Transmitted from mothers to sons -50% of child will be affected | back 9 Sex-linked recessive |
front 10 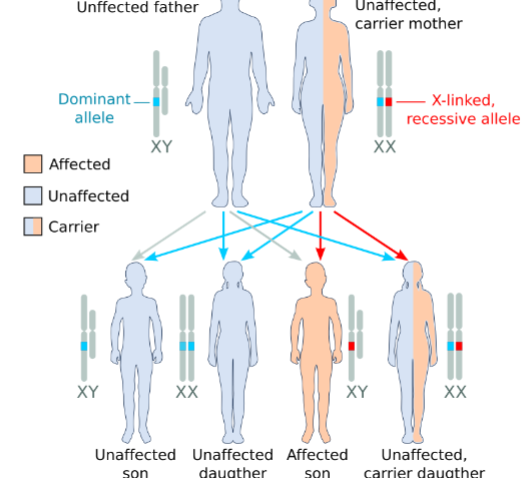 Example of__________ | back 10 Sex-linked recessive |
front 11 color blindness is___linked and affects _____more than _______ | back 11 -x -male -female |
front 12 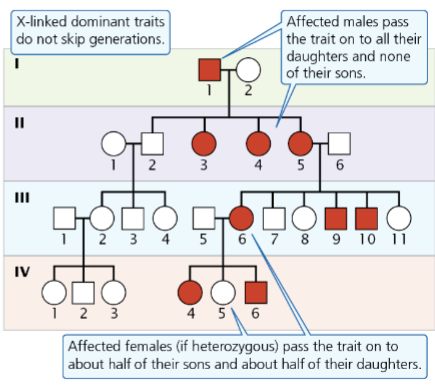 -affected offspring come from parents -males pass only to daughters -females pass to children of either sex -if dad is affected 100% of daughters are affected -if mom affected 100% of sons are affected | back 12 Sex-linked dominant |
front 13 What are examples of sex-linked dominant disorders? | back 13 -Alport syndrome -Coffin-Lowry syndrome -Fragile X syndrome -Incontinentia pigmenti -Vitamin D resistant rickets |
front 14 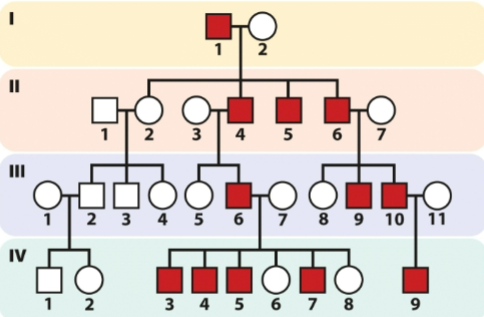 -only males have it -pass to all sons, none of daughters | back 14 Y-linked trait (dominant) |
front 15 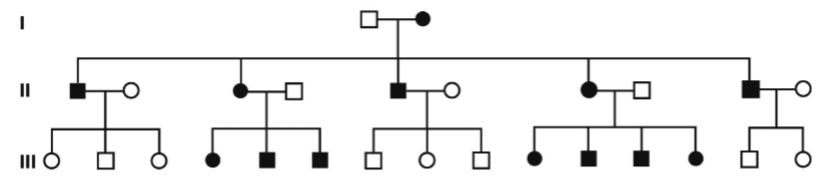 -Mothers always pass on -Fathers never pass on | back 15 Mitochondrial mutation |
front 16 What is the chance of their next child being affected? (a)= affected | back 16 (1/2) x (1/2)= (1/4) -gender x condition |
front 17 Look at slide 34 in Ch 6. pt 1 | back 17 DO IT NOW |
front 18 -non identical twins -2 zygotic eggs | back 18 Dizygotic twins |
front 19 -identical twins | back 19 Monozygotic twins |
front 20 -the trait shared by the members of a twin pair -genetically the same -environment may be similar -the increased rate is more likely impacted by genetic factors rather than environmental factors | back 20 concordant traits |
front 21 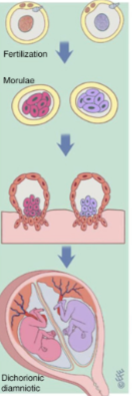 -separate embryotic sacks | back 21 dizygotic twinning |
front 22 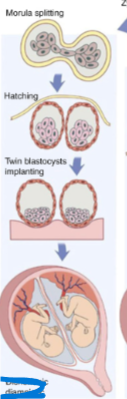 -comes from 1 zygote -early splitting | back 22 Dichorionic diamniotic (monozygotic twinning) |
front 23 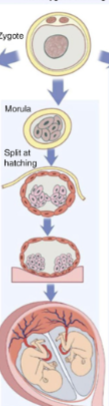 -late split | back 23 Monochorionic diamniotic (monozygotic twinning) |
front 24 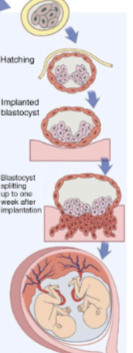 -very late split -same sack | back 24 Monochorionic monoamniotic (monozygotic twinning) |
front 25 Overweight biological parents tend to have_______children. There is no ___ _____ between the weight of children and that of their adoptive parents. | back 25 -overweight -consistent association |
front 26 This is an example of : | back 26 Environmental impact |
front 27 Genes on the same chromosomes tend to | back 27 get transmitted together |
front 28 certain combinations of alleles are passed more frequently than others | back 28 linkage |
front 29 Complete linkage leads to; ____% non recombinant gametes and progeny | back 29 100 |
front 30 the crossing over between ______ chromosomes can break linkage | back 30 homologous |
front 31 Alleles are passed in different combinations from how they were inherited | back 31 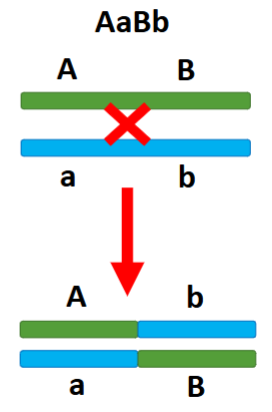 Recombination |
front 32 Recombination leads to: | back 32 -recombinant gametes and progeny |
front 33 ________ associated with physical change in chromosomes | back 33 recombinant phenotypes |
front 34 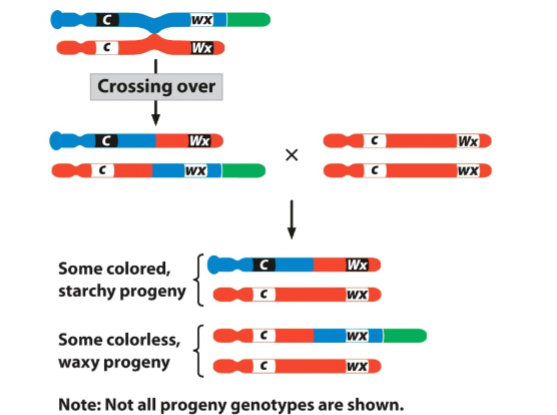 | back 34 recombinant diagram |
front 35 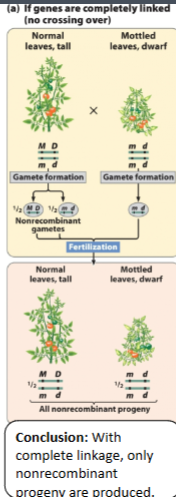 When a plant is normal with tall leaves and short with dwarf leaves, they are | back 35 nonrecombinant progeny |
front 36 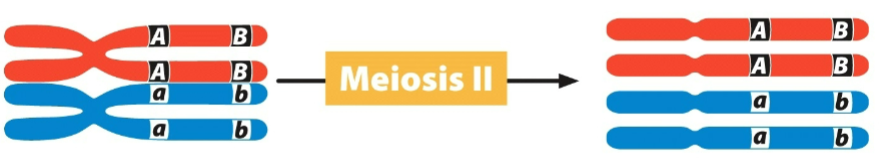 This is an example of | back 36 no crossing over |
front 37  This is an example of: which results is; | back 37 crossing over -2 nonrecombinant and 2 recombinant gametes |
front 38 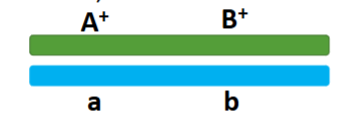 One chromosome contains both wild-type alleles, one chromosomes contains both mutant alleles | back 38 coupling (cis configuration) |
front 39  wild type allele and mutant alleles are found on the same chromosomes | back 39 repulsion (trans configuration) |
front 40 Usually 2 step process:\ 1. determine data are consistent with genes assorting independently (chi-squared test) 2. If the data is not consistent with independent assortment, this suggests genes are linked - determine how closely linked the genes are | back 40 determining genetic distance |
front 41 How to calculate recombination frequency | back 41 recombination freq.=(# of recombinant progeny/ total # of progeny)x100% |
front 42 No correlation between which alleles are transmitted=50% , recombinant gametes =50cM | back 42 independent assortment |
front 43 Two genes are linked if they are less than____ apart | back 43 50cM |
front 44 If the progeny resulted in: -66 -77 -28 -33 Which are recombinant, and what would the distance be? | back 44 -66 and 77 are the highest number= parental progeny -28 and 33 lowest numbers= recombinant progeny (28+33)/(66+77+28+33)=0.303 x100= 30.3 cM |
front 45 a crossover at one place on the chromosome reduces the likelihood of a second crossover | back 45 interference |
front 46 How to calculate amount of interference: coefficient of coincidence= observed DCO/expected DCO = | back 46 -((2)^3)/(total amount of flies x DCO freq.) -((2)^3)/12= 0.67 |
front 47 Distances based on crossover/linkage | back 47 genetic map |
front 48 Distances based on actual base-pair differences | back 48 physical map |
front 49 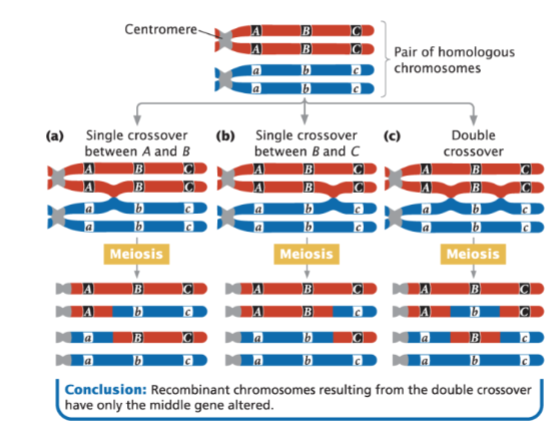 This is an example of | back 49 crossovers with three linked loci |
front 50 recombination distance are ______accurate the larger they get | back 50 less |
front 51 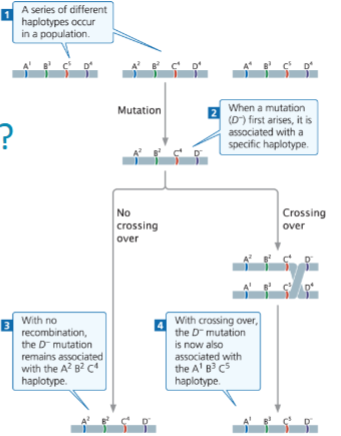 Consider the mutation and cross over | back 51 How halotype works |
front 52 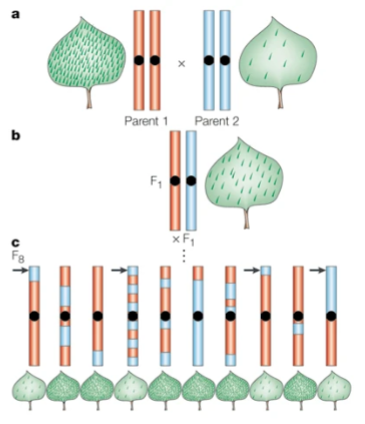 consider how when the top of loci is blue, it shows the blue parents trait | back 52 Quantitative trait locus mapping (QTL) |
front 53 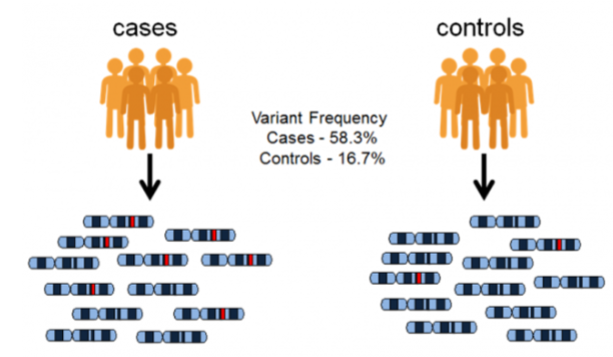 -controls have little to no genes of the condition -lower freq. of variant -variant linked to condition -cases have the condition | back 53 Genome-wide association studies (GWAS) |
front 54 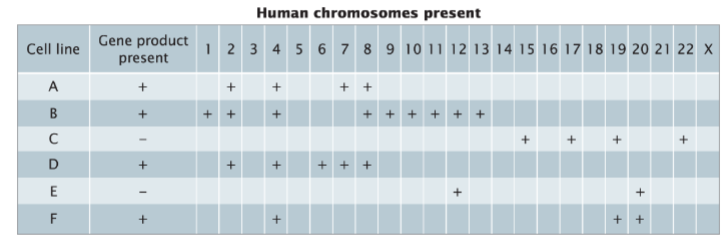 Used to assign a gene to a particular human chromosome -gene present (given) -Human chromosomes present ( find most similar to gene presented) | back 54 Somatic-cell hybridization |
front 55 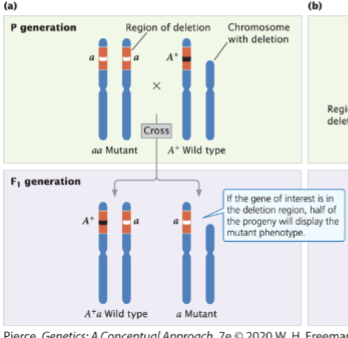 | back 55 displays the mutant phenotype |
front 56 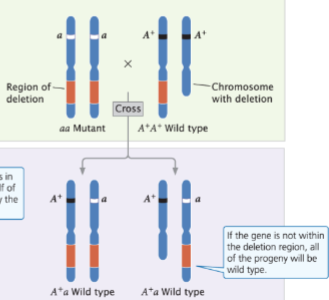 | back 56 displays the wild type |
front 57 -prepared from cells halted in metaphase(relaxed) - numbered based on size -Sex chromosomes -based on haploid # (23 chromosomes) | back 57 Karyotyping |
front 58 With sex chromosomes, the largest is the ___ chromosome and the short chromosome is the ___ chromosomes | back 58 -X -Y |
front 59 -moving, gaining, or losing pieces of a chromosome -2n=6 -duplication | back 59 rearrangement |
front 60 -changes in # of copies of an individual chromosome -2n+1=7 -trisomy | back 60 Aneuploidy |
front 61 -extra whole set of chromosomes -3n=9 -autotriploid | back 61 polyploidy |
front 62 Types of rearramgement: | back 62 -duplication -deletion -translocation -inversion |
front 63 Copied region of chromosomes | back 63 duplication |
front 64 deleted region of chromosomes | back 64 deletion |
front 65 inverting (flipping) region of chromosomes | back 65 inversion |
front 66 moving part of chromosome somewhere else | back 66 translocation |
front 67 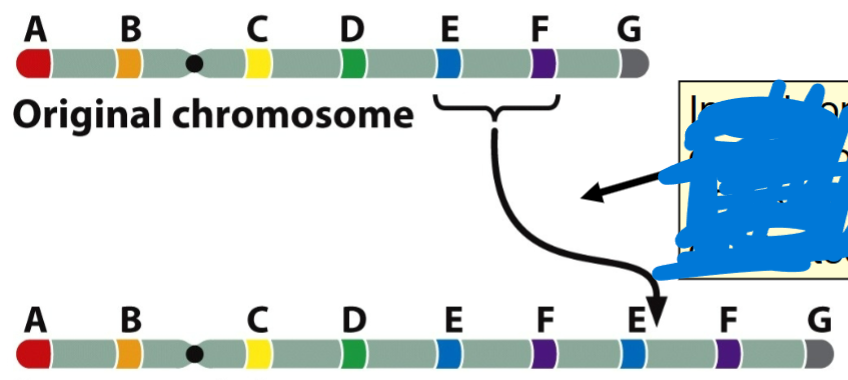 -bulge during pairing in meiosis 1 | back 67 Duplication (segment of chromosome is duplicated) |
front 68 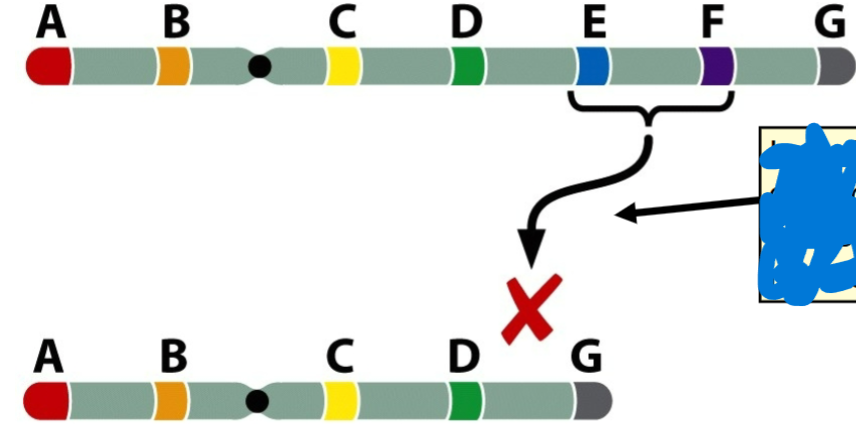 -formation of loop during pairing in prophase 1 | back 68 Deletion |
front 69 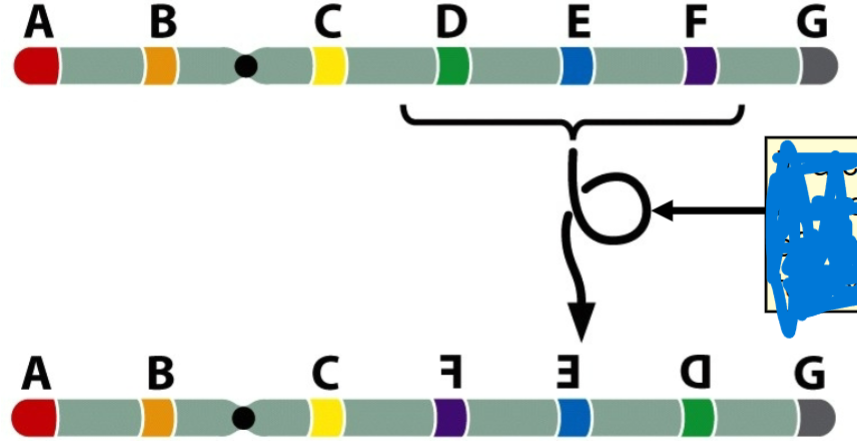 -turned 180 degrees | back 69 inversion |
front 70 Inversion that doesn't include the centromere | back 70 paracentric inversion |
front 71 inversion includes the centromere | back 71 pericentric inversion |
front 72 If a crossover were to occur in the inverted region, | back 72 it would result in a complication |
front 73 The packaging of tremendous amounts of genetic info. into the small space within a cell has been called | back 73 The ultimate storage problem |
front 74 Is overwound or underwound causing it to twist on itself | back 74 Supercoiling DNA |
front 75 -add two turns - occurs when DNA is overrotated; the helix twists on itself | back 75 Positive supercoil |
front 76 -removes 2 turns -occurs when DNA is underwound; the helix twists on itself in the opposite directions | back 76 Negative supercoiling |
front 77 What do these charactericstics describe? -less condensed -on chromosome arms -unique sequences -many genes -throughout S phase - often transcription - crossing over is common | back 77 Euchromatin |
front 78 What do these characteristics describe? -more condensed -At centromeres, telomeres, and other specific places - repeated sequence -Late S phase -infrequent transcription -crossing over is uncommon | back 78 Heterochromatin |
front 79 Regions of relaxed chromatin where active transcription is taking place | back 79 chromosome puffs |
front 80 Chromosome puffs on giant polytene chromosomes isolated from the __ ____ of larval Drosophila | back 80 -salivary glands |
front 81 Chromosome fragments that lack ______ are lost in mitosis | back 81 centromeres |
front 82 Proposes that mitochondria and chloroplasts in eukaryotic cells arose from bacteria | back 82 Endosymbiotic theory |
front 83 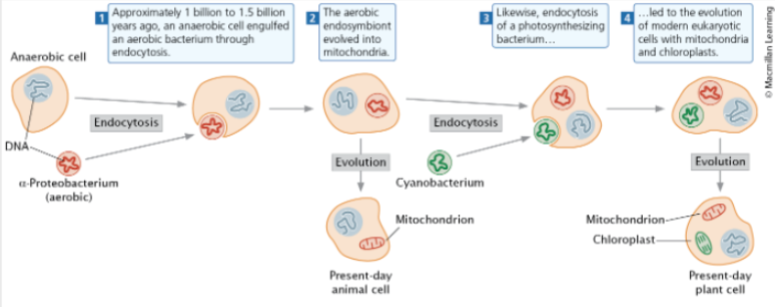 | back 83 endosymbiotic theory |
front 84 Organelles in a ______cell segregate randomly into the progeny cells. | back 84 heteroplasmic |
front 85 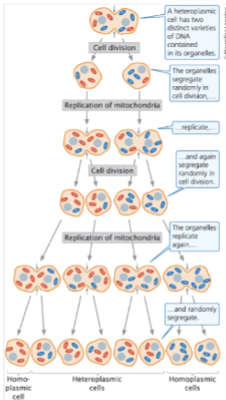 | back 85 understand it |
front 86 What is the importance of mutations? | back 86 -provides raw material for evolution -source of many diseases and disorders -look at whats wrong when we alter a gene |
front 87 -Do not get passed on to the offspring -passed to new cells through mitosis creating a clone of cells having mutant gene | back 87 Somatic mutations |
front 88 -reproductive cell mutations -passed on -meiosis and sexual reproduction allow mutation to be passed down to about half the members of next generation | back 88 germ-line mutation |
front 89 One letter is replaced by another | back 89 base substitution |
front 90 A to G & C to T (purines switch with purines) (pyrimidine switches with pyrimidine) | back 90 Transition |
front 91 A or G to C or T (purines switch with pyrimidines) (pyrimidine switch with purines) | back 91 Transversion |
front 92 Frameshift mutations are not_____by 3 | back 92 divisible |
front 93 in frame insertions and deletions are divisible by | back 93 3 |
front 94 Original sequence: GGG AGT GTA GAT CGT What would the sequence look like if a base substitution were to occur on the following nucleotide? | back 94 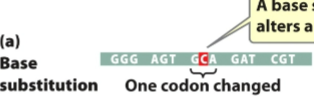 |
front 95 Original sequence: GGG AGT GCA GAT CGT What would the sequence look like if a base insertion were to occur on the following nucleotide? | back 95  |
front 96 Original sequence: GGG AGT GTT AGA TCG T What would the sequence look like if a base deletion were to occur on the following nucleotide? | back 96  |
front 97 Repetitive sequence can cause______ of strands during replication | back 97 slippage |
front 98 Dna polymerase adds | back 98 more repeats |
front 99 unequal cross over misaligns | back 99 homologous chromosomes |
front 100  The new codon encodes a different amino acid; there is a change in amino acid sequence | back 100 Missense mutation |
front 101 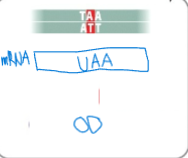 The new codon is a stop codon; there is a premature termination of translation | back 101 Nonsense mutation |
front 102 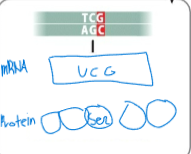 The new codon encodes the same amino acid; there is no change in amino acid sequence | back 102 Silent mutation |
front 103 a mutation that hides or suppresses the effect of another mutation Ex: wild type has red eyes (A+B+), and a mutation causes white eyes (A-B+). This allows for a fly with (A-B-) to have red eyes even if it has two mutant phenotypes. | back 103 Suppressor mutation |
front 104 Second mutation in the same gene "fixes" problem caused by first mutation | back 104 intragenic |
front 105 second mutation in a different gene. "fixes" the problem caused by the first mutation | back 105 Intergenic |
front 106 Purines and pyrimidine bases exist in different forms called | back 106 tautomers |
front 107 ____ _____ pairings can occur as a result of the flexibility in DNA structures EX: T and G binding (wobble) C and A protonated wobble | back 107 Nonstandard base |
front 108 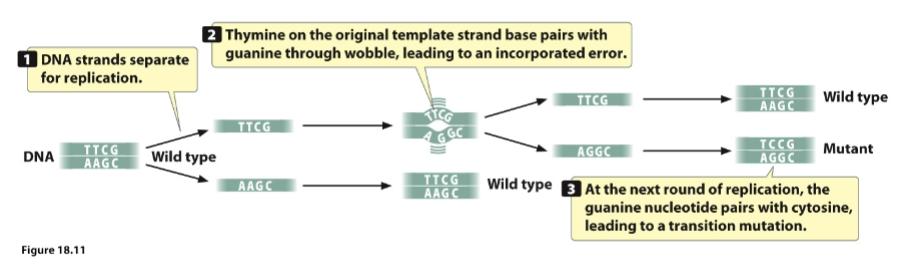 ____ base pairing leads to a replication error | back 108 Wobble |
front 109 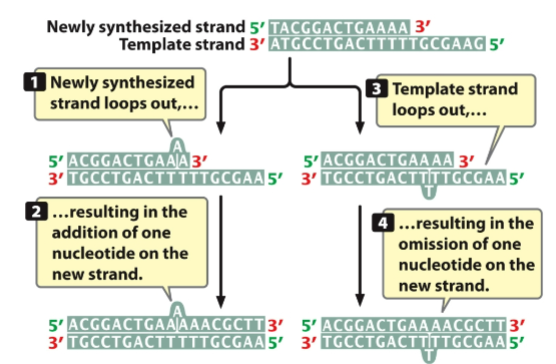 ____ and ____may result from strand slippage | back 109 -insertions -deletions |
front 110 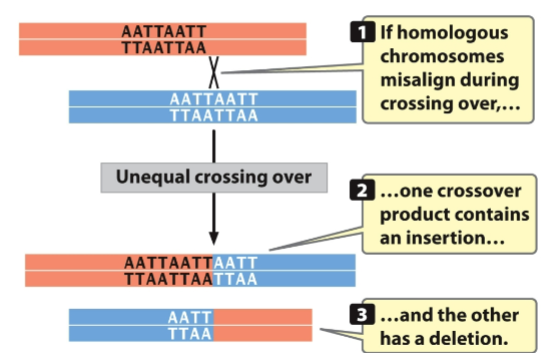 ____ _____ over produces insertions and deletions | back 110 unequal crossing over |
front 111 loss of purine | back 111 Depurination |
front 112 loss of an amino group | back 112 deamination |
front 113 Used to identify chemical mutagens | back 113 Ames test |
front 114 A___ can result in possible wobble mispairing | back 114 bulge |
front 115 Many incorrectly inserted nucleotides that escape proofreading are | back 115 corrected by mismatch repair |
front 116 Changes nucleotides back into their original structures | back 116 Direct repair |
front 117 A damaged base can be removed by______, and ______ cleaves the deoxyribose sugar. | back 117 -DNA glycosylase -AP endonuclease |
front 118 DNA _______ adds a new nucleotide and is then sealed by DNA _____ to restore the original sequence. | back 118 -polymerase -ligase |
front 119 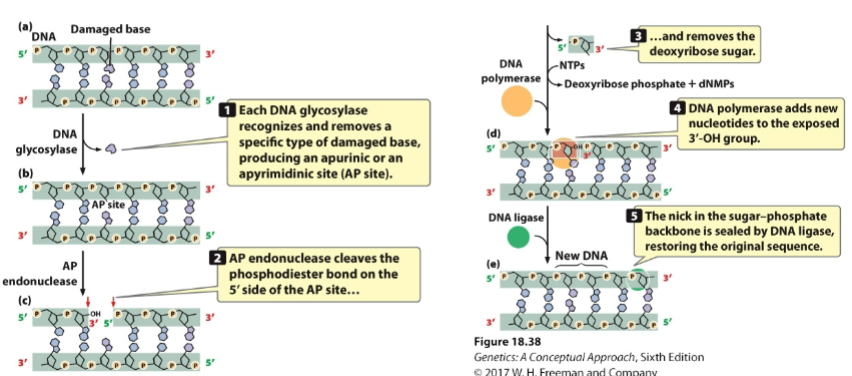 | back 119 base excision |
front 120 Removes bulky DNA lesions that distort double helix | back 120 Nucleotide-excision |
front 121 Can cause chromosomal rearrangement | back 121 transposons |
front 122 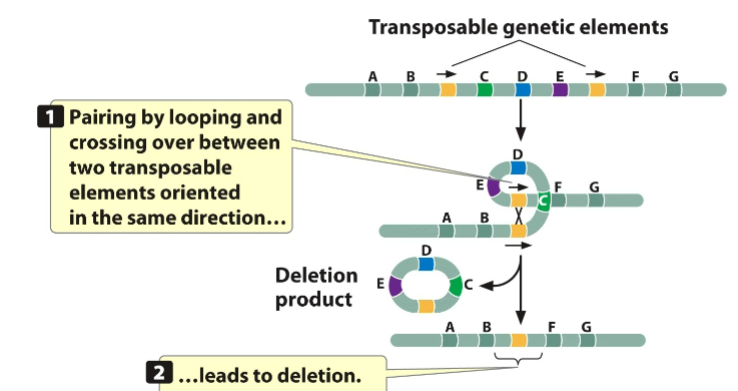 | back 122 example of transposons causing rearrangement |
front 123 How to calculate the frequencies of observed genotypes? -# of individuals with genotype divided by total # individuals f(AA)= f(Aa)= f(aa)= | back 123 -f(AA)= # of AA individuals/#of total individuals -f(Aa)= # of Aa individuals /# of total individuals -f(aa)=# of aa individuals /# of total individuals |
front 124 A population of 868 individuals has 482 AA individuals, 213 Aa individuals, and 173 aa individuals. What are the frequencies of each genotype? | back 124 f(AA)= 482/868=0.555 f(Aa)= 213/868=0.245 f(aa)= 173/868=0.199 |
front 125 How to calculate observed allelic frequencies -# copies of alleles divided by total # alleles f(A)= f(a)= | back 125 -f(A)=(2 x #AA individuals + #Aa individuals)/(2 x total # of individuals) -f(a)=(2 x #aa individuals + #Aa individuals)/(2 x total # of individuals) |
front 126 A population of 868 individuals has 482 AA people, 213 Aa people, and 173 aa people. What are the frequencies of each allele? | back 126 -f(A)= (2 x 482 +213)/(2 x 868)=0.678 -f(a)= (2 x 173 + 213)/(2 x 868)=0.322 |
front 127 What does p and q stand for in this equation? p+q=1 | back 127 -p: dominant -q: recessive |
front 128 equation 1 is used to determine___ p+q=1 equation 2 is used to determine___ p^2+2pq+q^2=1 | back 128 -frequencies of alleles in population -frequencies of genotypes in the population |
front 129 What are the Hardy-Weinberg Equilibrium Assumptions ? | back 129 -no selection -no mutation -no gene flow -random mating -infinite population size |
front 130 0.000625 Caucasians suffer from cystic fibrosis an autosomal recessive disease. (1)What is the frequency of the disease-causing allele in the population? (2) find value of p (3)What proportion of individuals are carriers? | back 130 -(1) q^2 =0.000625, q=√0.000625 =0.025 -(2) p+q=1 -> p=1-0.025= 0.975 -(3) 2pq= 2 x 0.975 x 0.025 = 0.049 , about 5% carry the cystic fibrosis mutation |
front 131 in cats, all-white is dominant over colors other than all white. In a population of 100 cats, 19 are all white. Assuming that the population is in HArdy-Weinburg equilibrium, what is the frequency of the all-white allele in this population? How many white cats would be expected to be genotype AA? Aa? | back 131 -white =19 out of 100, so 81 of the cats are not white(recessive) -81/100 = q=0.9 -p=1-0.9 = 0.1 are white and dominant |
front 132 Can cause severe changes in allelic frequencies and are more severe in smaller populations | back 132 Genetic drift |
front 133 Will alter the genotype frequencies, but not the allele frequencies of a population | back 133 nonrandom mating |
front 134 A tendency of like individuals to mate -creates more homozygotes then expected -flies with same eye color mate | back 134 Positive assortative mating |
front 135 A tendency of unlike individuals to mate. -creates more heterozygotes than expected -fly with red eyes mates with white eyes | back 135 Negative assortative mating |
front 136 -increases the likelihood of getting two copies of the same allele from the same ancestor results in more homozygotes | back 136 inbreeding |
front 137 a measure of how likely it is that two alleles are identified by descent | back 137 inbreading coefficient (F) |
front 138 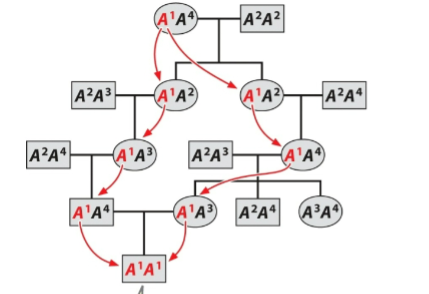 What is this an example of? | back 138 Alleles identical by descent |
front 139 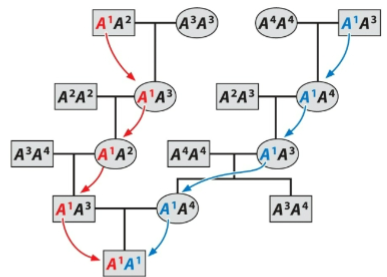 What is this an example of? | back 139 alleles identical by state |
front 140 The more inbreeding the less | back 140 heterozygotes |
front 141 Caused by accumulation of harmful recessive alleles | back 141 Inbreeding depression |
front 142 What equations are needed when calculating with inbreeding factors? | back 142 f(AA)= p^2 + Fpq f(Aa)=2pq-2Fpq f(aa)= q^2 +Fpq |
front 143 What will the genotype frequencies be after 1 generation of random mating, followed by 1 generation of inbreeding between siblings?(F=0.25) p=0.6 q=0.4 | back 143 Fpq= 0.25 x 0.6 x 0.4=0.06 F(AA)= p^2 =0.6^2=0.36 +0.06= 0.42 F(Aa)= 2pq= 2 x 0.6 x 0.4=0.48 -(2 x 0.06) =0.36 F(aa)=q^2=0.4^2=0.16 + 0.06 = 0.22 |
front 144 -Doesn't change allele frequencies by itself but changes distribution of who has them -more homozygous -recessive alleles that are not good will more likely show | back 144 non-random mating |
front 145 Individuals from one population entering another population | back 145 migration |
front 146 The amount of change in allelic frequency due to migration between populations depends on | back 146 the difference in allelic frequency and the extent of migration |
front 147 changes in allele frequency due to different levels of fitness (ability to survive and reproduce) | back 147 Natural selection |
front 148 One allele is always more fit than the other -the more "fit" allele increases in frequency | back 148 Directional selection |
front 149 The heterozygote is more fit than either homozygotes -stable equilibrium reached and both alleles remain in population | back 149 Overdominance (heterozygote advantage) |
front 150 Heterozygote is less fit than either homozygotes -unstable equilibrium reached- one allele will eventually win | back 150 Under dominance (heterozygote disadvantage) |
front 151  What will be the allele frequncies at the time reproduction beings ? | back 151 1. divide by the most fit number in this case 180 AA=180/180=1 Aa=162/180= 0.9 aa= 36/180= 0.2 2. A=(180+180+heterozygote)/(2x total number of individuals) (180 + 180 +162) / (162+180+36) x 2= 0.69 a= (36 +36+ 162) / (756)= 0.31 |
front 152 w11<w12>w22 is an example of what? | back 152 -overdominance |
front 153 w11<w12<w22 is an example of ? | back 153 Directional selection against incompletely dominant allele A1 |
front 154 w11>w12>w22 is an example of? | back 154 Directional selection against incompletely dominant allele A2 |
front 155 w11=w12<w22 is an example of? | back 155 directional selection against dominant allele A1 |
front 156 w11=w12>w22 is an example of | back 156 directional selection against recessive allele A2 |
front 157 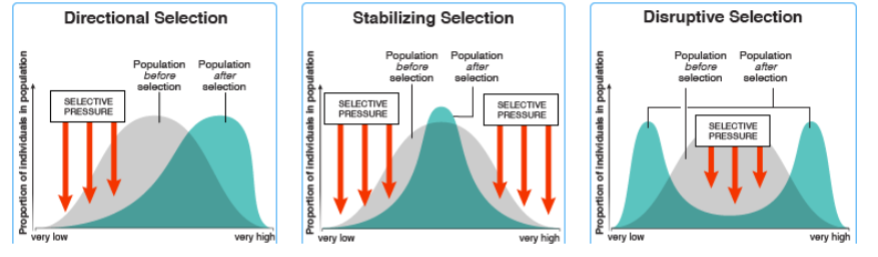 | back 157 consider this when looking at distributions |