Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
genetics test 7-9
front 1 1) The process by which Pneumococcus transfers DNA between living type RII and heat-killed type SIII cells is known as
| back 1 transformation |
front 2 2) Avery, Macleod, and McCarty expanded on Griffith's experiment to prove that DNA is the hereditary molecule required for transformation. What treatment of the heat-killed SIII bacteria extract resulted in the mouse LIVING?
| back 2 Destruction of type SIII DNA with DNase |
front 3 3) In the Hershey-Chase experiment, bacteriophages were produced in either 32P-containing or 35S-containing medium. Where were these isotopes eventually detected when the radioactively-labeled bacteriophages were introduced to a fresh bacterial culture?
particles.
| back 3 D) The 32P was associated with the bacterial cells and 35S was associated with the phage particles. |
front 4 4) Which of the following are classified as pyrimidines?
| back 4 E) thymine and cytosine |
front 5 5) What type of bond is formed between the hydroxyl group of one nucleotide and the phosphate group of an adjacent nucleotide, forming the sugar-phosphate backbone of DNA?
| back 5 C) phosphodiester bond |
front 6 6) What is the difference between a nucleotide and a nucleoside?
| back 6 C) A nucleoside with a phosphate ester linked to the sugar is a nucleotide. |
front 7 7) What is the difference between a ribonucleic acid and a deoxyribonucleic acid?
| back 7 A) Deoxyribonucleic acids have a 2'H instead of the 2'OH found in ribonucleic acids. |
front 8 8) What types of bonds are formed between complementary DNA bases?
| back 8 B) hydrogen bonds |
front 9 9) If there is 24% guanine in a DNA molecule, then there is cytosine.
| back 9 C) 24% |
front 10 10) In a single strand of DNA, what fraction of the nucleotides in a molecule are cytosine and thymine (% C and % T added together)?
| back 10 E) It depends on the DNA sequence |
front 11 11) If complementary DNA strands were arranged in a parallel manner, what would you expect to see?
so the strands could be more easily pulled apart.
an antiparallel manner.
would make the helix stable but not uniform in width.
of the two DNA strands would not be stable. | back 11 E) Some regions of the two strands may form atypical hydrogen bonds, but the overall structure of the two DNA strands would not be stable. |
front 12 12) In their famous experiment, which of the following would Meselson and Stahl have observed after one cycle of replication in 14N medium if DNA replication were CONSERVATIVE?
| back 12 A) An equal number of DNA molecules containing two 15N-DNA strands and DNA molecules containing two 14N-DNA strands |
front 13 13) A portion of one strand of DNA has the sequence 5′ AATGGCTTA 3′. If this strand is used as a template for DNA replication, which of the following correctly depicts the sequence of the newly synthesized strand in the direction in which it will be synthesized?
| back 13 C) 3′ TTACCGAAT 5′ |
front 14 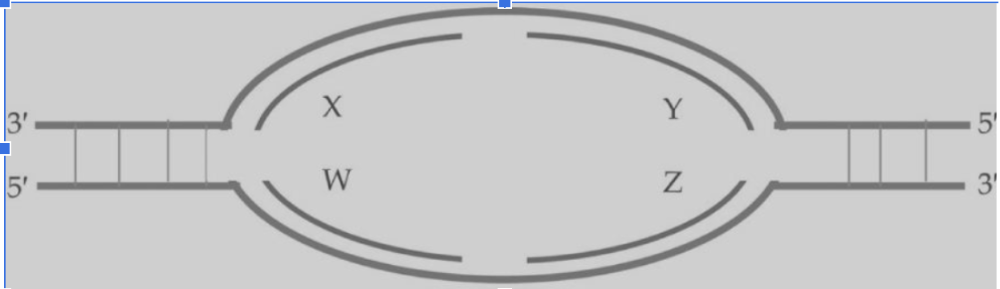 14) Based on the following replication bubble, which of these statements is true?
| back 14 C) W and Y are leading strands, X and Z are lagging strands |
front 15 15) The following represents a DNA strand in the process of replication. The bottom sequence is that of DNA strand with polarity indicated and the top sequence represents the RNA primer. GGGGCCUUU 5′ AAATCCCCGGAAACTAAAC 3′ Which of the following will be the first DNA nucleotide added to the primer?
| back 15 A) A |
front 16 16) What is one difference between DNA replication in bacteria versus eukaryotes?
origin of replication.
replicated in one direction.
while bacteria have only one origin of replication and replicate uni-directionally.
replicated in one direction. | back 16 A) Eukaryotic chromosomes have many origins of replication, while bacteria have only one origin of replication. |
front 17 17) What is the DNA replication fork?
lagging strand.
| back 17 B) It is the site where the DNA helix opens to two single DNA strands. |
front 18 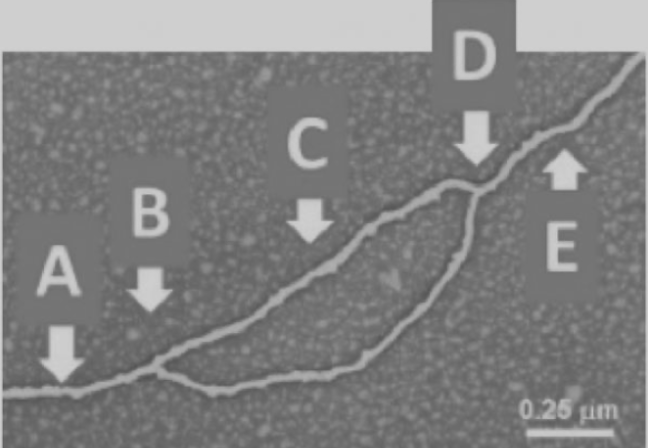 18) Which arrow(s) point(s) to the location of helicase in the diagram shown?
| back 18 B and D |
front 19 19) Why would DNA synthesis occur in both directions from an origin of replication?
| back 19 D) Replication-associated proteins bind at two replication forks. |
front 20 20) Why do origins of replication in various bacteria have conserved DNA consensus sequences?
initiation proteins.
regions.
change it.
origin as well.
why they are conserved as consensus sequences. | back 20 B) Origins need to be recognized by replication initiation proteins to open up their AT-rich regions. |
front 21 21) If Single-Stranded Binding protein (SSB) is NOT present during DNA replication, what would you expect to see?
DNA replication cannot proceed.
| back 21 C) SSB prevents reannealing of the separated strands, so strands would quickly reanneal and DNA replication cannot proceed. |
front 22 22) DNA helicase inhibitors are well studied as potential drug targets. What would you expect to see if DNA helicase activity is inhibited?
and DNA replication cannot proceed.
unwound and strands will not separate.
separation will not occur.
| back 22 C) Helicase catalyzes ATP hydrolysis and DNA strands separation, so the helix cannot be unwound and strands will not separate. |
front 23 23) What is required for DNA polymerase to initiate DNA strand synthesis?
| back 23 C) a short RNA primer synthesized by the enzyme primase |
front 24 24) Why are leading and lagging strand primers removed rather than joined with Okazaki fragments?
Polymerase. | back 24 E) They contain nucleotides with 2'OH groups, and are targeted for excision by DNA Polymerase. |
front 25 25) You identify a cell in which DNA polymerase III is functional, but it seems to exhibit extremely low processivity. This is likely a defect in what structure?
| back 25 E) the sliding clamp |
front 26 26) The extraordinary accuracy of the DNA polymerase III enzyme lies in its ability to "proofread" newly synthesized DNA, a function of the enzyme's _____.
| back 26 C) 3′-to-5′ exonuclease activity |
front 27 27) What is the purpose of the 3'-to-5' exonuclease activity of DNA Polymerase?
synthesis in the 5' to 3' direction.
| back 27 B) Remove mismatched nucleotides in the newly synthesized strand. |
front 28 28) Where would you expect to find telomerase activity?
| back 28 C) At the end of a chromosome in a cancerous eukaryotic body cell. |
front 29 29) Why are telomeres problematic for eukaryotic chromosome replication?
| back 29 E) Removal of the lagging strand primer leaves a gap in the one of the strand's DNA sequences. |
front 30 30) Which of the following would you find in a Sanger sequencing reaction but not in a polymerase chain reaction?
| back 30 E) ddNTPs |
front 31 31) Which functional groups have been altered in a ddNTP compared to a dNTP?
| back 31 E) The ddNTPs have a 2′ H and a 3′ H, while dNTPs have a 2′ H and a 3′ OH. |
front 32 32) Which of the following temperature cycles would you expect to see in a standard polymerase chain reaction (from denaturation to annealing to extension)?
| back 32 C) 95° → 55° → 72° |
front 33 33) Why are next generation DNA sequencing technologies known as sequencing-by-synthesis?
| back 33 B) Incorporated nucleotides are determined while they are being added to a growing DNA strand. |
front 34 1) What are two distinguishing features of RNA?
E) RNA contains a methylated form of thymine. | back 34 B |
front 35 2) Which type of research technique was used to track newly synthesized RNA within a eukaryotic cell?
| back 35 C |
front 36 3) Prokaryotes and eukaryotes produce which of the following types of RNA?
| back 36 B |
front 37 4) You wish to create a mutation that prevents access of RNA polymerase to the gene. Which region of a gene would you mutate?
E) coding region | back 37 B |
front 38 5) A gene has acquired a mutation in which the protein product has 50 additional amino acids at the end. Which region of the gene was likely mutated?
| back 38 B |
front 39 6) What is the role of a promoter region of a gene?
| back 39 B |
front 40 7) Which region(s) of a gene are NOT found within the mRNA
transcript? D) promoter and stop codon | back 40 E |
front 41 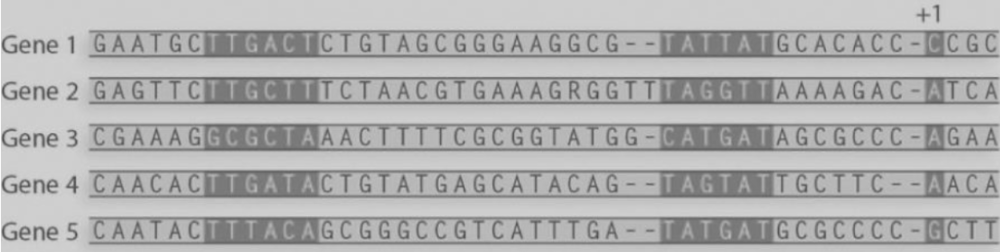 8) What is the consensus sequence for the Pribnow box from these sequences?
E) TATGAT | back 41 E |
front 42  9) What is the -35 consensus sequence for the following sequences?
| back 42 A |
front 43 10) In a given bacterium, transcription of housekeeping genes is normal, but genes involved in nitrogen metabolism, stress, and chemotaxis are disrupted. Which sigma subunit is INTACT?
| back 43 D |
front 44 11) What is the significance of the open complex when the RNA Polymerase binds the DNA? A) The RNA Polymerase binds the single-stranded template strand in
its active site. B) The RNA Polymerase binds the single-stranded
coding strand in its active site. | back 44 A |
front 45 12) Why does rho-dependent transcriptional termination in bacteria require the rho protein? A) RNA Polymerase stalls at various sites in the gene and rho helps
push RNA Polymerase to the end of the gene. | back 45 C |
front 46 13) You want to design a drug that prevents transcription of
eukaryotic mRNAs but does not affect 13) transcription of other RNAs.
What enzyme would you target?
| back 46 B |
front 47 14) What is the type of each eukaryotic protein that primarily transcribes mRNA, tRNA, and rRNA, 14) respectively?
| back 47 A |
front 48 15) What must eukaryotes do to initiate transcription of a gene?
D) Recruit the transcription factors and RNA Polymerase that compose
the pre-initiation complex. | back 48 D |
front 49 16) Which assay allows you to identify the exact location of the
protein-binding sequence within a promoter?
E) pulse-chase assay | back 49 C |
front 50 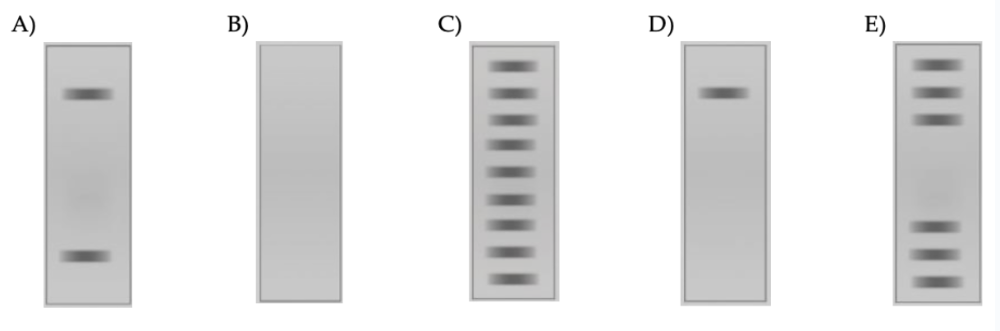 17) Following a DNA footprint protection assay, which banding pattern would you expect to see if you identify a potential promoter region? | back 50 E |
front 51 18) RNA polymerase I transcribes which tandem-repeat genes in nucleoli? A) tRNA | back 51 B |
front 52 19) Which of the following is part of a DNA molecule?
E) promoter | back 52 E |
front 53 20) Which of the following statements accurately describes tRNA?
E) tRNAs are a variety of lengths and fold into a variety of shapes. | back 53 C |
front 54 21) Which of the following are present in your liver cells? A) promoters of genes expressed in the kidney | back 54 E |
front 55 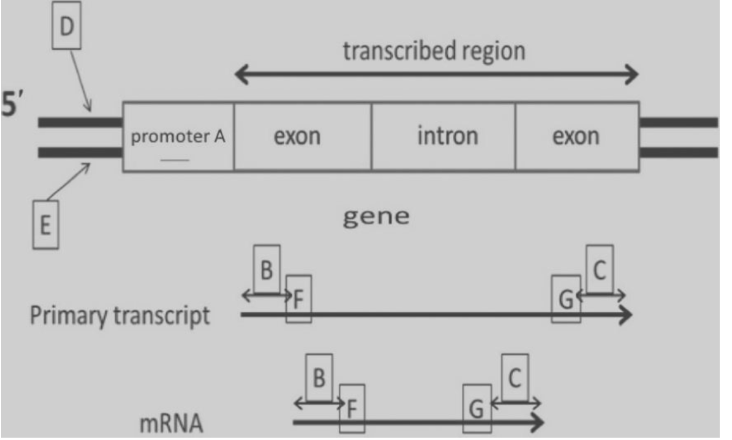 22) If transcription of this gene occurs from left to right in the accompanying diagram, which DNA strand is the CODING (non-template) strand?
| back 55 D |
front 56  23) Using the accompanying diagram, which of the following corresponds to the 3' untranslated region?
| back 56 C |
front 57 24) Which enzyme is required to initiate 5′ capping of eukaryotic mRNA transcripts by removing the terminal phosphate group?
D) ribozyme | back 57 C |
front 58 25) What defines the end of a eukaryotic gene?
E) Presence of a polyadenylation signal sequence leads to cleavage of the pre-mRNA. | back 58 E |
front 59 26) A cell has a defect in polyadenylation of mRNA. The RNA transcripts encoding which type of protein would NOT be affected by this defect because they are not polyadenylated?
| back 59 B |
front 60 27) What is the general name for the components of the spliceosome, which removes introns from mRNAs?
| back 60 B |
front 61 28) What is the purpose of alternative splicing in eukaryotic cells?
D) Improve the efficiency of transcription and translation. | back 61 A |
front 62 29) The rat α-tropomyosin (α-Tm) gene produces nine different mature mRNA proteins from a single gene using which three "alternative" mechanisms?
| back 62 A |
front 63 30) What are catalytically active RNAs that can activate processes such as self-splicing?
E) ribozymes | back 63 E |
front 64 31) In humans, the 30S pre-RNA transcript yields three rRNA segments
following transcription 31) and RNA cleavage. Which RNA transcripts
are generated by the 30S pre-RNA transcript in
| back 64 A |
front 65 32) One type of RNA editing involves inserting uracils into edited mRNA with the assistance of which 32) type of specialized RNA?
E) guide RNA | back 65 E |
front 66 1) What features of proteins does two-dimensional gel electrophoresis exploit in order to separate proteins?
| back 66 B |
front 67 2) The Shine-Dalgarno sequence in bacteria .
| back 67 E |
front 68 3) During translation initiation in bacteria, the amino acid on the initiator tRNA is .
| back 68 D |
front 69 4) Identification of ribosomal proteins involves two-dimensional gel electrophoresis, which separates the proteins on the basis of .
| back 69 B |
front 70 5) How does the eukaryotic initiation complex locate the correct start codon?
the start AUG.
sequence.
Sequence. | back 70 A |
front 71 6) What is the cellular location of the stages of translation in bacteria and eukaryotes?
| back 71 B |
front 72 7) How does the eukaryotic ribosomal small subunit recognize the start codon on the mRNA?
codon.
already bound to the AUG. | back 72 A |
front 73 8) A tRNA in the P site of the ribosome will enter the site after translocation of the ribosome. A. 3' B. E C. A D. initiation E. 5’ | back 73 B |
front 74 9) A portion of an mRNA attached to a ribosome reads: 5′ GACCAUUUUUGA 3′ If a tRNA with a Phenylalanine amino acid attached is in the P site of the ribosome, an empty tRNA present in the E site that delivered which amino acid?
| back 74 D |
front 75 10) A portion of mRNA attached to a ribosome reads: 5′ GACCAUUUUUGA 3′ In the polypeptide produced, what amino acid will be attached to the amino group of the histidine encoded by this mRNA?
| back 75 D |
front 76 11) What would you expect to find bound to the stop codon at the A site?
| back 76 D |
front 77 12) ) What is necessary for a eukaryotic RNA to be recognized and bound by the small subunit of the ribosome?
| back 77 C |
front 78 13) Elongation factors translocate the ribosome in the 3′ direction by a distance of .
| back 78 B |
front 79 14) A polycistronic mRNA contains multiple .
| back 79 D |
front 80 15) Why are eukaryotic mRNAs not polycistronic, unlike some bacterial transcripts?
ribosome can bind to a Shine-Delgarno sequence anywhere in the mRNA.
in initiating translation.
same mRNA.
sufficient information to encode additional polypeptides. | back 80 A |
front 81 16) What does it mean for two codons to be synonymous?
| back 81 E |
front 82 17) What result would you expect if a mutation eliminates one of the four arms of a tRNA?
| back 82 C |
front 83 18) How many different aminoacyl-tRNA synthetases can be found in a given organism's cells?
| back 83 D |
front 84 19) If a tRNA anticodon were mutated such that it no longer performed wobble, what would be the effect on encoded proteins?
| back 84 A |
front 85 20) A mutagen has introduced a frame-shift mutation by adding one nucleotide base. Which of the following could be a reversion mutation for this particular mutant?
| back 85 D |
front 86 21) Which of these choices represents one possible corresponding mRNA sequence that can be transcribed from the following DNA template? 5′ - CTGTATCCTAGCACCCAAATCGCATTAGGAC - 3′
| back 86 D |
front 87 22) Given the following mRNA sequence, what is the amino acid sequence for the corresponding polyp 5′ - AUG CGA UUU GGG UGC UAG - 3′
| back 87 E |
front 88 23) Given the following mRNA sequence, which of the following mRNAs would encode a protein with a different sequence of amino acids? 5′ - AUG CAG UUA GCG UGC UAG - 3′
| back 88 B |
front 89 24) Which mRNA below would code for a premature stop codon from the following amino acid sequence N—Met-Gln-Leu-Arg-Cys—C
| back 89 D |
front 90 25) How might a single base INSERTION into the second codon of the coding sequence of a gene affect the amino acid sequence of a protein encoded by the gene?
| back 90 A |
front 91 26) You have identified a bacterial protein that has retained the starting fMet in its protein sequence. Which of the following is likely true of this protein?
functional proteins.
| back 91 C |
front 92 27) How is a tRNA able to recognize its proper mRNA codon?
site conformation to allow tRNA binding.
to the mRNA.
bonding. | back 92 D |
front 93 28) If the first nucleotide in a codon is mutated to a different nucleotide, what would be the effect on the encoded protein?
| back 93 D |
front 94 29) In the unlikely event that a tRNA has been charged with the wrong amino acid, which high-fidelity enzyme most likely caused the incorrect charging?
| back 94 A |
front 95 30) If the genetic code were overlapping, how many complete codons would the following sequence en before encountering a stop codon? 5′ - AUGCGAUUAAAGUGC - 3′
| back 95 B |
front 96 1) Studies of gene mutation frequencies have shown that A) mutation are common and adaptive | back 96 E) mutations are rare, and genomes are generally stable |
front 97 2) Which type of mutation is possible due to the redundant nature of the genetic code?
| back 97 E) silent |
front 98 3) Given the sequence of triplet codons: 5′-TAC AAA ATA CAG CGG-3′, which of these sequences represents a nonsense mutation?
E) 5′-TAG AAA ATA CAG CGG-3′ | back 98 5′-TAG AAA ATA CAG CGG-3′ |
front 99 4) Given the sequence of triplet codons: 5′-TAC AAA ATA CAG CGG-3′, which of these sequences 4) represents a missense mutation?
| back 99 B) 5′-TAC AAA ATA CAC CGG-3′ |
front 100 5) Which of the following is able to cause a change in a reading frame? A) transversion B) transition | back 100 D) deletion |
front 101 6) What type of mutation is seen here? Mutation: 5′-TAC AAG ATA CAG CGG-3′
| back 101 D) transition |
front 102 7) A mutant DNA polymerase that increases the frequency of strand
slippage would increase the frequency of which type of mutation? | back 102 E) triplet-repeat expansion |
front 103 8) Which type of mutation converts a nucleotide to an alternative structure with the same composition but a slightly different placement of rare, less stable hydrogen bonds that cause base-pair mismatch?
E) transversion | back 103 A) tautomeric shift |
front 104 9) The fluctuation test allowed Luria and Delbruck to conclude that A) mutations are common and thus allow natural selection B) mutations occur in the absence of environmental challenges C) mutations occur at random in response to environmental challenges D) chemical mutagens are present even in drinking water E) none of the above | back 104
|
front 105 10) You would like to induce a transversion mutation into a sequence
of DNA. Which type of chemical mutagen would give you the best chance
of inducing the correct mutation without causing B) base analog | back 105 D) oxidative agent |
front 106 11) Transposons can integrate into the promoters of genes, what is the most likely outcome of such an event? A) amino acid substitution B) deamination C) point mutation D) altered gene expression E) frame shift | back 106 D) altered gene expression |
front 107 12) You have conducted an Ames test on a given compound. Which of the following would be classified as a positive result on the Ames test? A) his- strain grows on an his+ B) his+ strain grows on either an his- or an his+ plate. C) his- strain grows on an his- D) his+ strain grows on an his- E) his+ strain grows on an his+ plate. | back 107 C) his- strain grows on an his- |
front 108 13) A strain of E. coli is unable to use the UV repair pathway
because of an apparent absence of DNA helicase activity. Which UV
repair gene is likely mutated in this strain? B) uvr-A C) uvr-C D) uvr-D E) pol I | back 108 D) uvr-D |
front 109 14) What phenotype would you expect to see in a strain of E. coli with a mutation in the phr gene? A) decrease in UV-induced mutations B) decrease in reversion of base-pair substitution mutations C) increase in UV-induced mutations D) increase in the methylation of nucleotide bases E) decrease in the methylation of nucleotide bases | back 109 C) increase in UV-induced mutations |
front 110 15) Which pathway is affected by an inherited mutation in the ATM gene? A) nucleotide excision repair pathway | back 110 C) p53 repair pathway |
front 111 16) Which of these statements best describes gene expression in a damaged cell? A) In a damaged cell, p53 is high and BAX transcription is active,
so Bcl-2 is repressed and apoptosis is induced. E) In a damaged cell, p53 is high and BAX transcription is inactive, so Bcl-2 is activated and apoptosis is induced. | back 111 A) In a damaged cell, p53 is high and BAX transcription is active, so
Bcl-2 is repressed and |
front 112 17) Which of the following statements is true of non-homologous end joining (NHEJ)? 17) A) it is both error-prone and is a double-strand repair
pathway | back 112 A) it is both error-prone and is a double-strand repair pathway |
front 113 18) In eukaryotes, homologous recombination is initiated by A) Rad51 generating double-stranded DNA breaks B) ATM signaling double-stranded DNA breaks C) p53 generating single-stranded DNA breaks D) RecA, RecB, and RecC generating single-stranded DNA breaks E) Spo11 generating double-stranded DNA breaks | back 113 E) Spo11 generating double-stranded DNA breaks |
front 114 19) If one Holliday junction region is resolved by an NS cut and the other by an EW cut, the resulting chromosomes . A) are recombinant and carry the combinations A1 and B1 or A2 and B2 B) are nonrecombinant and carry the combinations A1 and B2 or A2 and B1 C) are nonrecombinant and carry the combinations A1 and B1 or A2 and B2 D) areA1 andA2 or B1 andB2 E) are recombinant and carry the combinations A1 and B2 or A2 and B1 | back 114 E) are recombinant and carry the combinations A1 and B2 or A2 and B1 |
front 115 20) Which enzyme is required to mobilize transposons of any type? A) RNA helicase B) terminal inverted repeats C) reverse transcriptase | back 115 D) transposase |