Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
Non-Mendl Ch 5, 16 Connect Assignment
front 1 If the expression of genes in the offspring directly influences their traits, the genes are transmissible from generation to generation, the genes segregate during gamete formation, and the genes of the trait segregate independently, then the genes follow ______ inheritance patterns. Multiple choice question. Mendelian non-Mendelian | back 1 Mendelian |
front 2 In an inheritance pattern called ____ ____, the genotype of the mother directly determines the phenotype of her offspring for a given gene. | back 2 maternal effect |
front 3 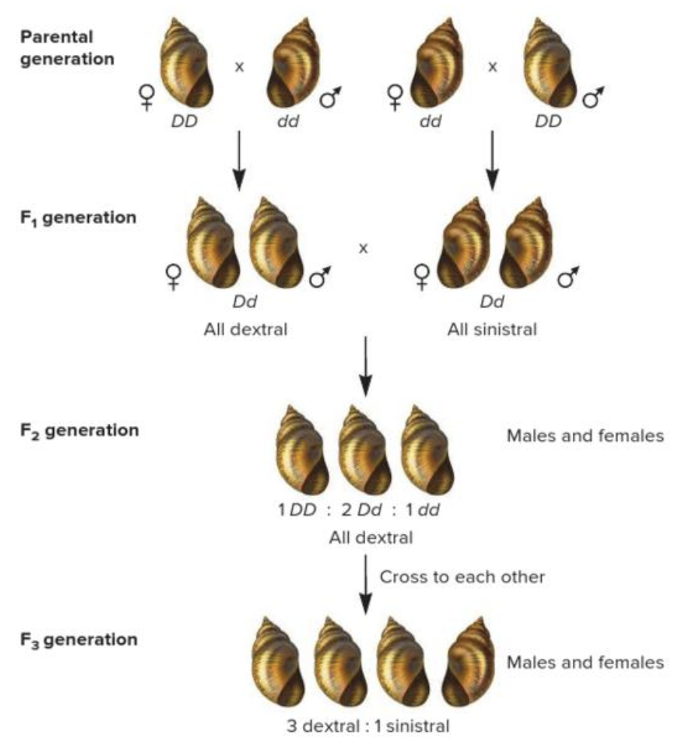 What accounts for the phenotype of the offspring in the F1 generation shown in the image? Multiple choice question. Paternal effect X-linked inheritance Codominance Sex influenced effect Maternal effect | back 3 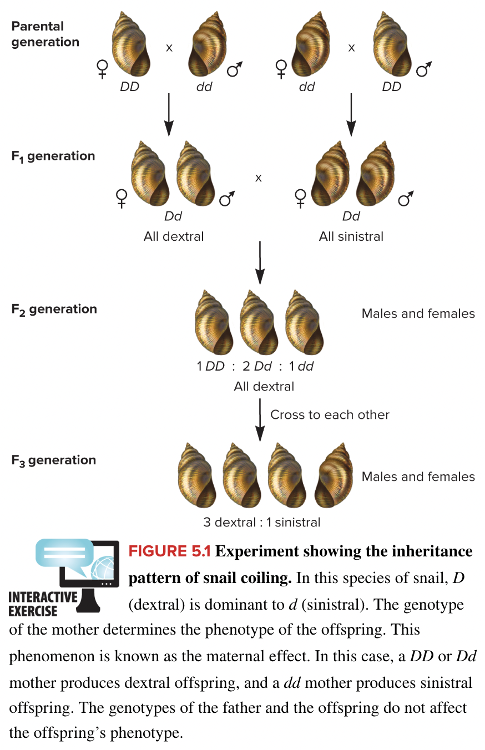 Maternal effect Reason: Dextral is dominant to sinistral. The genotype of the mother determines the phenotype of the offspring. This is known as maternal effect. In this case, a DD or Dd mother will produce dextral offspring and a dd mother produces sinistral offspring. The genotypes of the father and offspring do not affect the offspring's phenotype. |
front 4 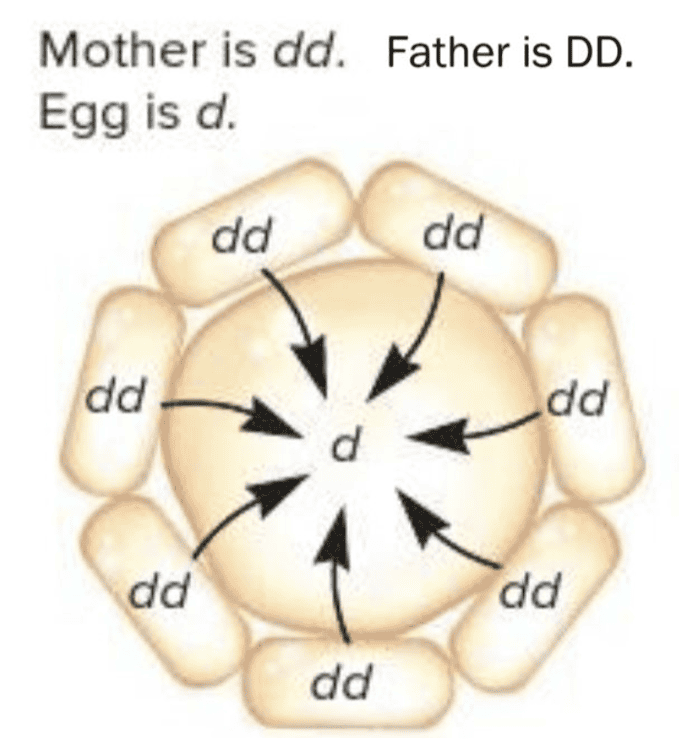 Snail shell coiling is due to a maternal effect gene with two alleles. The dominant allele D causes dextral (right-handed) shell coiling while the recessive allele d leads to sinistral (left-handed) coiling. The gene product is transferred from nurse cells into the egg as shown in the diagram for a homozygous (dd) mother. Suppose this egg is fertilized by a gamete from a dextral (DD) male. What are the expected phenotypes of the offspring? Multiple choice question. dextral sinistral both sinistral and dextral | back 4 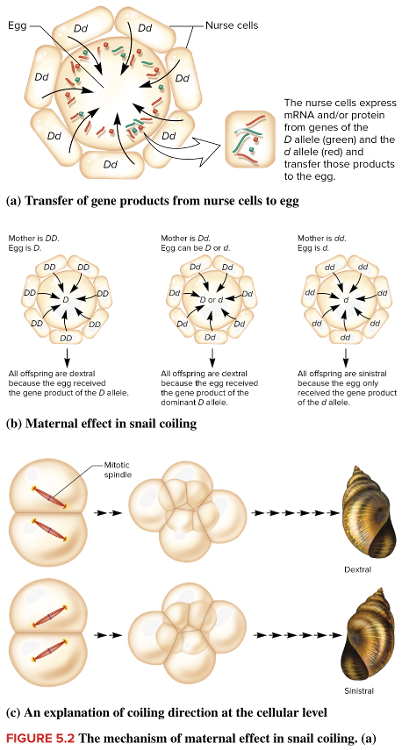 sinistral |
front 5 Within the two-to four-cell snail embryo, which factor is responsible for the origin of dextral and sinistral shell coiling? Multiple choice question. Orientation of the mitotic spindle Organization of proteins encoded by maternal genes Arrangement of proteins in the plasma membrane Location of vesicles produced by the Golgi apparatus | back 5 Orientation of the mitotic spindle |
front 6 Select all that apply Select all of these that are true of genes that follow a Mendelian inheritance pattern. Multiple select question. The genes are influenced by expression of maternal genes in the egg. The genes are transmissible from generation to generation. The genes segregate during gamete formation. Expression of genes in the offspring directly influences their traits. The genes segregate independently. | back 6 The genes are transmissible from generation to generation. The genes segregate during gamete formation. Expression of genes in the offspring directly influences their traits. The genes segregate independently. |
front 7 True or false: In Drosophila, proper anteroposterior and dorsoventral development requires a separate set of maternal effect gene products accumulating in the appropriate area of the embryo. | back 7 True Reason: Studies in Drosophila, mice and humans have identified several maternal effect genes that are required for proper development. |
front 8 What type of inheritance accounts for the genotype of the mother being the direct determining factor in the phenotype of the offspring? Multiple choice question. X-linked inheritance Sex influenced effect Codominance Paternal effect Maternal effect | back 8 Maternal effect |
front 9 A type of change in which a temporary alteration to a chromosome modifies gene expression but is not permanent over many generations is called ____ change. | back 9 epigenetic |
front 10 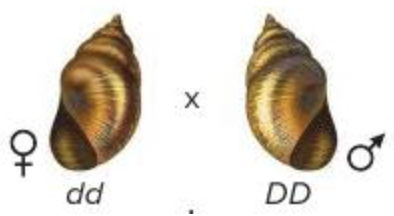 In the snail Lymnaea peregra, shell coiling is regulated by a maternal effect gene, and dextral is dominant to sinistral. What is the expected phenotype of the offspring for the cross shown in the image? Multiple choice question. 50% sinistral : 50% dextral 100% sinistral 75% sinistral : 25% dextral 100% dextral | back 10 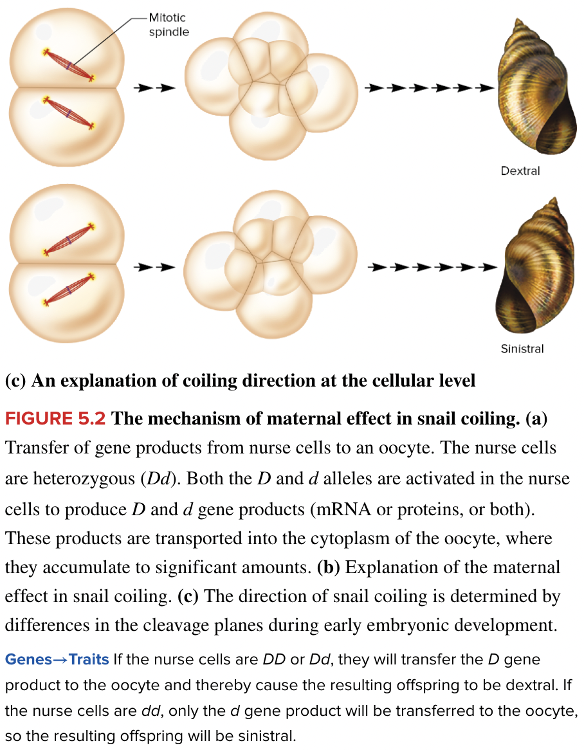 100% sinistral Reason: Dextral is dominant to sinistral. The genotype of the mother determines the phenotype of the offspring. This is known as maternal effect. In this case, a DD or Dd mother will produce dextral offspring and a dd mother produces sinistral offspring. The genotypes of the father and offspring do not affect the offspring's phenotype. |
front 11 Despite the fact that human males are XY and human females are XX, they have equal expression of many genes on their sex chromosomes. This phenomenon is called | back 11 dosage compensation [Blank 1: dosage, dosage compensation, X-chromosome, X, or dose Blank 2: compensation or inactivation] |
front 12 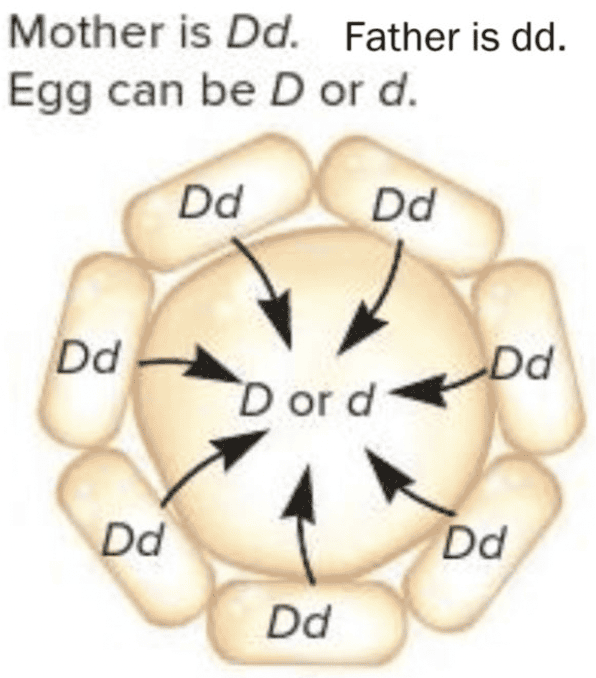 Snail shell coiling is due to a maternal effect gene with two alleles. The dominant allele D causes dextral (right-handed) shell coiling while the recessive allele d leads to sinistral (left-handed) coiling. The gene product is transferred from nurse cells into the egg as shown in the diagram for a heterozygous (Dd) mother. Suppose this egg is fertilized by a gamete from a sinistral (dd) male. What is the expected phenotype of the offspring? Multiple choice question. dextral if the egg has the D genotype sinistral if the egg has the d genotype sinistral for both the D and d genotypes in the egg dextral for both the D and d genotypes in the egg | back 12 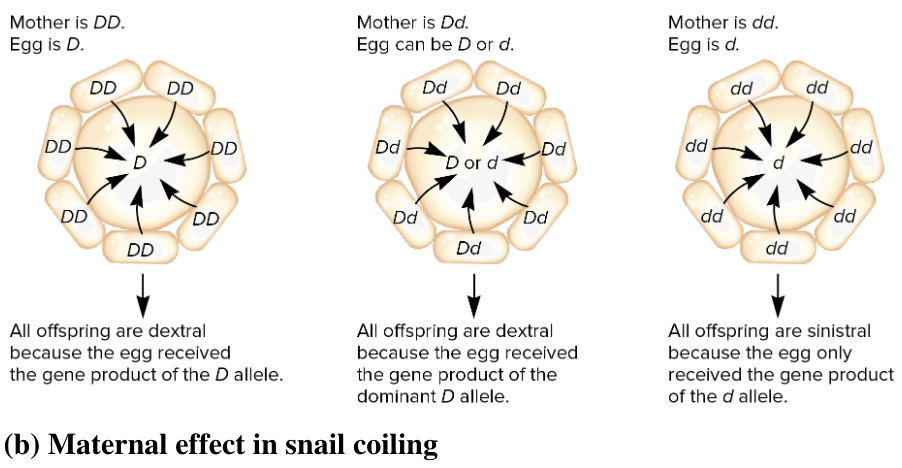 dextral for both the D and d genotypes in the egg |
front 13 In somatic cells of human females one of the two X chromosomes is randomly turned off. What is this mechanism called? Multiple choice question. Maternal effect inheritance Endosymbiosis Paternal leakage X-chromosome inactivation | back 13 X-chromosome inactivation |
front 14 The origin of dextral and sinistral coiling of snail shells can be attributed to the orientation of the ____ ____ at the two- to four-cell stage of embryonic development. | back 14 mitotic spindle |
front 15 In birds males are ZZ and females are ZW. Studies have shown that male birds express twice as much gene product of some Z-linked genes. What does this suggest? Multiple choice question. Z-linked genes are not dosage compensated in birds. Female birds decrease Z gene expression by 50%. One of the Z chromosomes is randomly inactivated in birds. | back 15 Z-linked genes are not dosage compensated in birds. Reason: The level of Z-linked gene expression should be equal if inactivation occurred. |
front 16 Select all that apply Select all of these that are influenced by maternal effect gene products. Multiple select question. Body axis orientation Fertilization Cleavage pattern Oogenesis Cell division | back 16 Body axis orientation Cleavage pattern Cell division |
front 17 Which species achieves dosage compensation by increasing the expression of genes on the X chromosome in males by twofold? Multiple choice question. Drosophila melanogaster Humans Marsupial mammals Caenorhabditis elegans | back 17 Drosophila melanogaster Reason: In humans, either the maternal or paternal X chromosome is randomly inactivated in somatic cells of females. Reason: Marsupial mammals achieve dosage compensation by inactivating the paternally derived X chromosome in all somatic cells of a female. Reason: In Caenorhabditis elegans, the level of expression on both X chromosomes in hermaphrodites is decreased to 50% levels compared with males. |
front 18 True or false: Epigenetic changes to a chromosome are permanent modifications that remain over the lifetime and later generations of the organism. | back 18 False Reason: Epigenetic changes to a chromosome modify the chromosome during oogenesis, spermatogenesis or embryogenesis, but are not permanent over the course of many generations. |
front 19 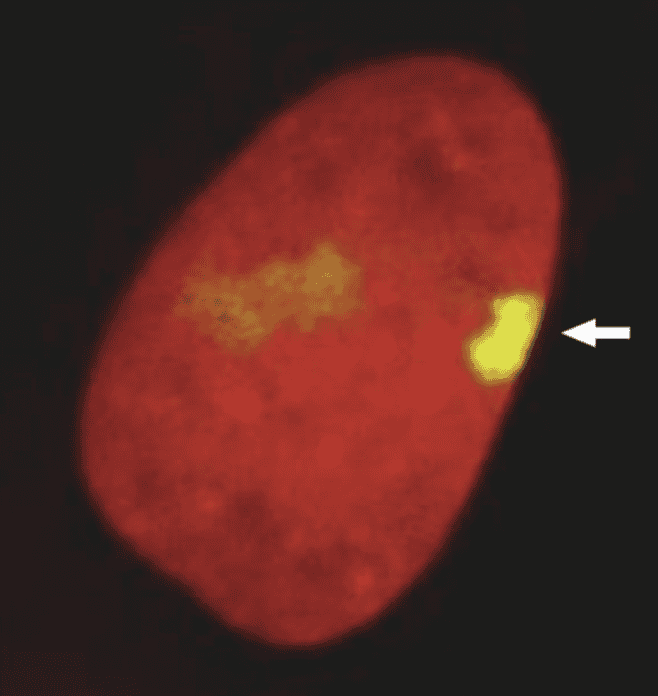 The inactivated X chromosome in somatic cells of mammals, like the one shown by the arrow in this image, is called a | back 19  Barr body |
front 20 What accounts for equal expression of genes on the sex chromosomes despite the fact that males and females have an unequal complement of sex chromosomes? Multiple choice question. Sex influenced inheritance Sex limited inheritance Maternal effect Dosage compensation | back 20 Dosage compensation |
front 21  The coat color of the calico cat shown in the image is best explained by ______. Multiple choice question. maternal effect paternal effect codominance the Lyon hypothesis | back 21  the Lyon hypothesis Reason: X chromosome inactivation is a random process that occurs early on in development. This is what accounts for the variegated phenotype. |
front 22 In humans with a typical constitution of chromosomes, X-chromosome inactivation induces dosage compensation by randomly inactivating one of the X chromosomes in the somatic cells of ______. Multiple choice question. both males and females males females | back 22 females |
front 23 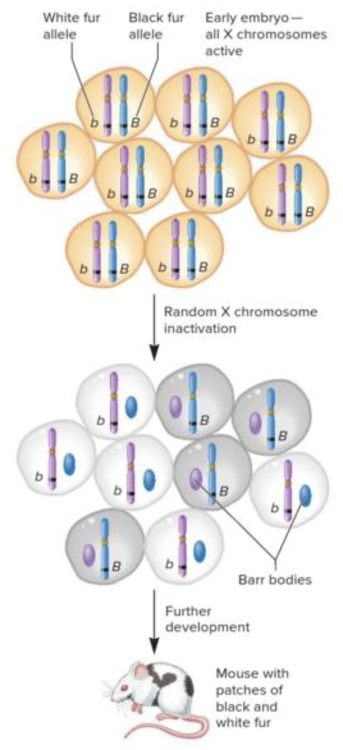 The image is depicting the mechanism of X-chromosome inactivation, called the ______. Multiple choice question. genomic imprinting maternal effect theory of independent assortment Lyon hypothesis | back 23 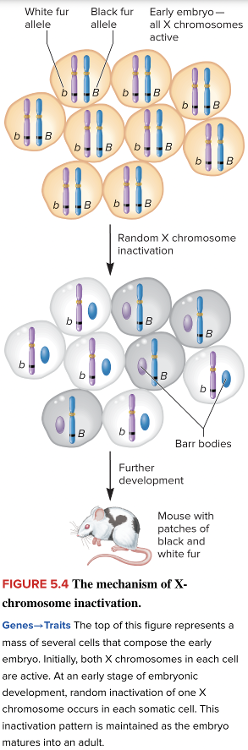 Lyon hypothesis |
front 24 True or false: Like X chromosomes in human females, one of the Z chromosomes in female birds is randomly inactivated in somatic cells. | back 24 False Reason: Studies suggest that some Z-linked genes may be dosage compensated, but many are not. Male birds have two similar sex chromosomes. |
front 25 Which statement best describes how X chromosomes are inactivated in female mammals? Multiple choice question. The same X is inactivated in each gamete. The same X is inactivated in all cells at an early stage of embryonic development. One X is randomly inactivated in each cell at an early stage of embryonic development. One X is randomly inactivated in each gamete. | back 25 One X is randomly inactivated in each cell at an early stage of embryonic development. Reason: The inactivation process is random, either of the two X chromosomes in a given cell can be inactivated. |
front 26 Which species achieves dosage compensation by inactivating the paternally derived X chromosome in all somatic cells of a female? Multiple choice question. Drosophila melanogaster Humans Caenorhabditis elegans Marsupial mammals | back 26 Marsupial mammals Reason: In Drosophila melanogaster, the level of expression of genes on the X chromosome in males is increased twofold. Reason: In humans, either the maternal or paternal X chromosome is randomly inactivated in somatic cells of females. Reason: In Caenorhabditis elegans, the level of expression on both X chromosomes in hermaphrodites is decreased to 50% levels compared with males. |
front 27 In the experiment carried out by Davidson, Nitowsky, and Childs, which outcome would support the Lyon hypothesis? Multiple choice question. All nine clones expressed the slow G-6-PD protein. All nine clones expressed either the fast or slow G-6-PD protein but not both. All nine clones expressed both the fast and slow G-6-PD proteins. All nine clones expressed the fast G-6-PD protein. | back 27 All nine clones expressed either the fast or slow G-6-PD protein but not both. Reason: One of the genes would be inactivated so both protein forms would not be expressed. |
front 28 What is a Barr body? Multiple choice question. An inactivated X chromosome in mammalian somatic cells An inactivated autosome in mammalian somatic cells An inactivated Y chromosome in mammalian somatic cells | back 28 An inactivated X chromosome in mammalian somatic cells |
front 29 Population of cells that are all derived from a single cell are called | back 29 clones |
front 30 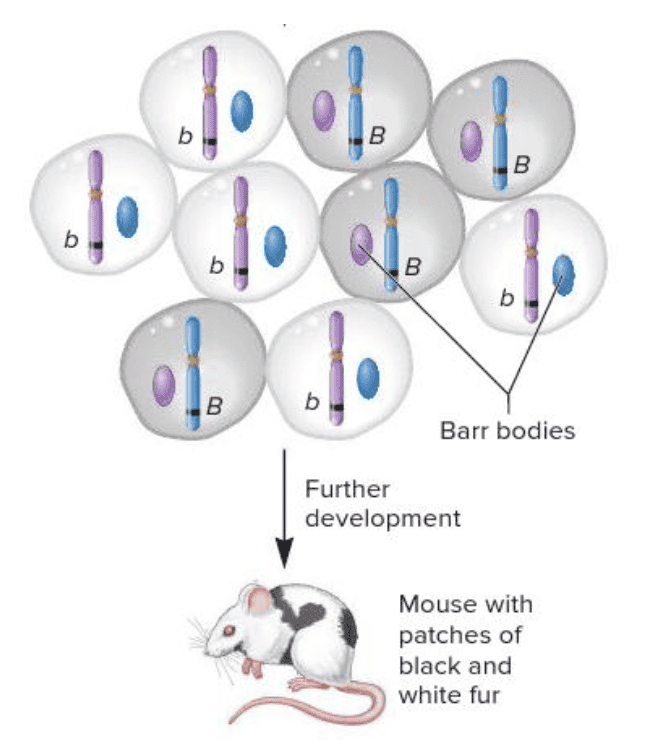 As shown in the image, X chromosomes are randomly inactivated during embryonic development. This is also called ______. Multiple choice question. codominance the Lyon hypothesis paternal effect maternal effect | back 30  the Lyon hypothesis |
front 31 A male with Klinefelter syndrome (XXY) would have how many Barr bodies in his cells? Multiple choice question. One Two Three None | back 31 One Reason: The number of Barr bodies is equal to the number of X chromosomes minus one. |
front 32 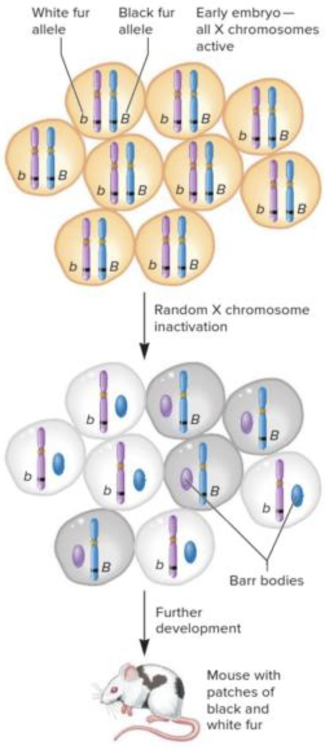 Fill in the blank question. The mechanism of X-chromosome inactivation (shown in the figure) is known as the ____ hypothesis. | back 32  Lyon |
front 33 The short region on the X chromosome that plays an important role in X chromosome inactivation is the ____. (Use an abbreviation.) | back 33 Xic |
front 34 What happens to the inactivated X chromosome in female mammals when cells divide? Multiple choice question. The inactivated X chromosome is passed along to all future somatic cells. The inactivated X chromosome decondenses and one of the two X chromosomes is randomly inactivated in each somatic daughter cell. The inactivated X chromosome stays inactivated in both somatic cells and gametes. | back 34 The inactivated X chromosome is passed along to all future somatic cells. Reason: The pattern of inactivation is passed along during somatic cell division. Reason: Inactivation occurs in somatic cells. |
front 35 Select all that apply Select all of these that are phases of X chromosome inactivation. Multiple select question. Nucleation Spreading Maintenance Termination Elongation | back 35 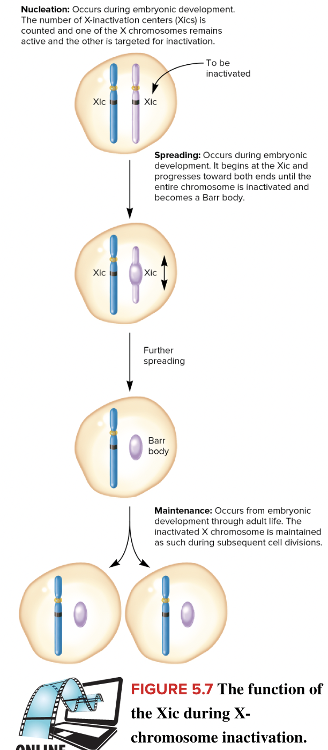 Nucleation Spreading Maintenance |
front 36 For the experiment testing the Lyon hypothesis at the cellular level carried out by Davidson, Nitowsky, and Childs, the individual who donated the cells must have been a ______ for the gene of interest. Multiple choice question. male homozygous female heterozygous male heterozygous female homozygous | back 36 female heterozygous Reason: Only heterozygotes can show which gene is inactivated, either for the slow or fast form of the protein. |
front 37 Select all that apply Which phases of X chromosome inactivation occur only during embryonic development? Multiple select question. Maintenance Nucleation Spreading | back 37  Nucleation Spreading Reason: Maintenance occurs from embryonic development throughout life to preserve the state of X chromosome inactivation. |
front 38 Which of these best describes a clone? Multiple choice question. A population of cells derived from a single cell Oocytes fertilized by the same sperm donor Gametes produced by meiosis | back 38 A population of cells derived from a single cell |
front 39 Select all that apply A person who has somatic cells with two Barr bodies could have which of the following genotypes? Multiple select question. XXY XXX XXXY XO XYY | back 39 XXX XXXY |
front 40 True or false: All genes on the inactivated X chromosome remain inactivated throughout life in a somatic cell of an adult female. | back 40 False Reason: Some genes on the inactivated X chromosome are expressed in the somatic cells of adult female mammals. |
front 41 What is the primary role of the X-inactivation center (Xic)? Multiple choice question. Expression of genes on the X chromosome X-chromosome inactivation Expression of female secondary characteristics | back 41 X-chromosome inactivation N: Xic = X-inactivation center |
front 42 What are the three phases of X chromosome inactivation? Multiple choice question. Initiation, elongation, and termination Denaturation, annealing and elongation Nucleation, spreading, and maintenance Replication, transcription and translation | back 42 Nucleation, spreading, and maintenance |
front 43 During which phase of X chromosome inactivation is one of the X chromosomes chosen to be inactivated? Multiple choice question. Spreading Maintenance Nucleation | back 43 Nucleation Reason: The X chromosome is targeted for inactivation during the initiation phase. The actual inactivation occurs during the spreading phase. |
front 44 What is genomic imprinting? Multiple choice question. A form of inheritance in which organellar genetic material influences gene expression A form of epigenetic inheritance in which a segment of DNA is marked in a way that it alters gene expression The expression of genes based on inheritance through the cytoplasm of the egg The inactivation of an X chromosome by Xist gene RNAs | back 44 A form of epigenetic inheritance in which a segment of DNA is marked in a way that it alters gene expression Reason: Genomic imprinting refers to a situation in which a segment of DNA is marked before fertilization, and that mark is retained and recognized throughout the life of the organism inheriting the marked DNA. |
front 45 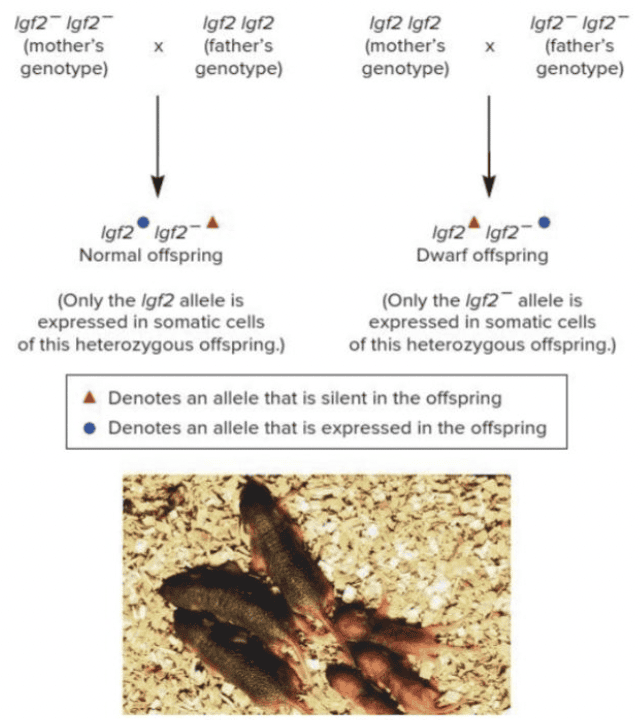 The mice in the figure are heterozygous, Igf2 Igf2 -, yet they have different phenotypes due to genomic imprinting. What is this phenomenon called? Multiple choice question. Codominance Monoallelic expression Variable expression Incomplete dominance | back 45  Monoallelic expression |
front 46 Which genes on the X chromosome are able to escape the effects of X chromosome inactivation in female mammalian cells? Multiple choice question. FoxP3 gene SRY gene Pseudoautosomal genes Paternal effect genes AZF1 gene | back 46 Pseudoautosomal genes |
front 47 In the case of the Igf2 gene, which copy is expressed in the offspring? Multiple choice question. The allele from the father Both alleles It can be either the allele from the mother or the father, depending on which is imprinted. The allele from the mother | back 47 The allele from the father Reason: Imprinting only occurs for the paternal Igf2 allele. |
front 48 Patterns of imprinting are maintained in what type of cells throughout development? Multiple choice question. Somatic Germ-line | back 48 Somatic |
front 49 Select all that apply Select all of the true statements regarding genomic imprinting. Multiple select question. The marking of alleles cannot be altered from generation to generation. Genomic imprinting is permanent in somatic cells. The marking of alleles can be altered from generation to generation. Genomic imprinting is temporary in somatic cells. | back 49 Genomic imprinting is permanent in somatic cells. The marking of alleles can be altered from generation to generation. Reason: Imprinting can be changed in the production of sex cells. |
front 50 In a form of epigenetic inheritance called ____ ____, a segment of nuclear DNA is marked in a non-permanent way that alters gene expression throughout the life of the individual. | back 50 genomic imprinting |
front 51 At the molecular level, one way genomic imprinting is regulated is by DNA | back 51 methylation |
front 52 Even though offspring receive two alleles, one maternal and one paternal, during genomic imprinting only one allele is expressed. What is this phenomenon called? Multiple choice question. Codominance Monoallelic expression Variable expression Incomplete dominance | back 52 Monoallelic expression |
front 53 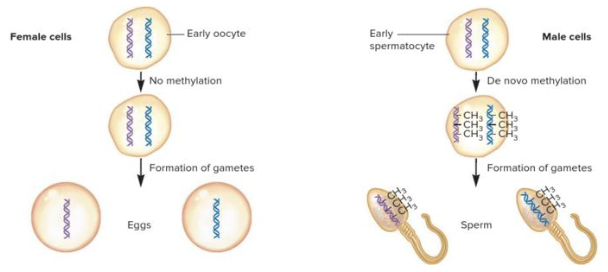 Based on the information in the figure,will the paternal or maternal chromosome be active in the resulting zygote? Multiple choice question. Maternal Paternal | back 53 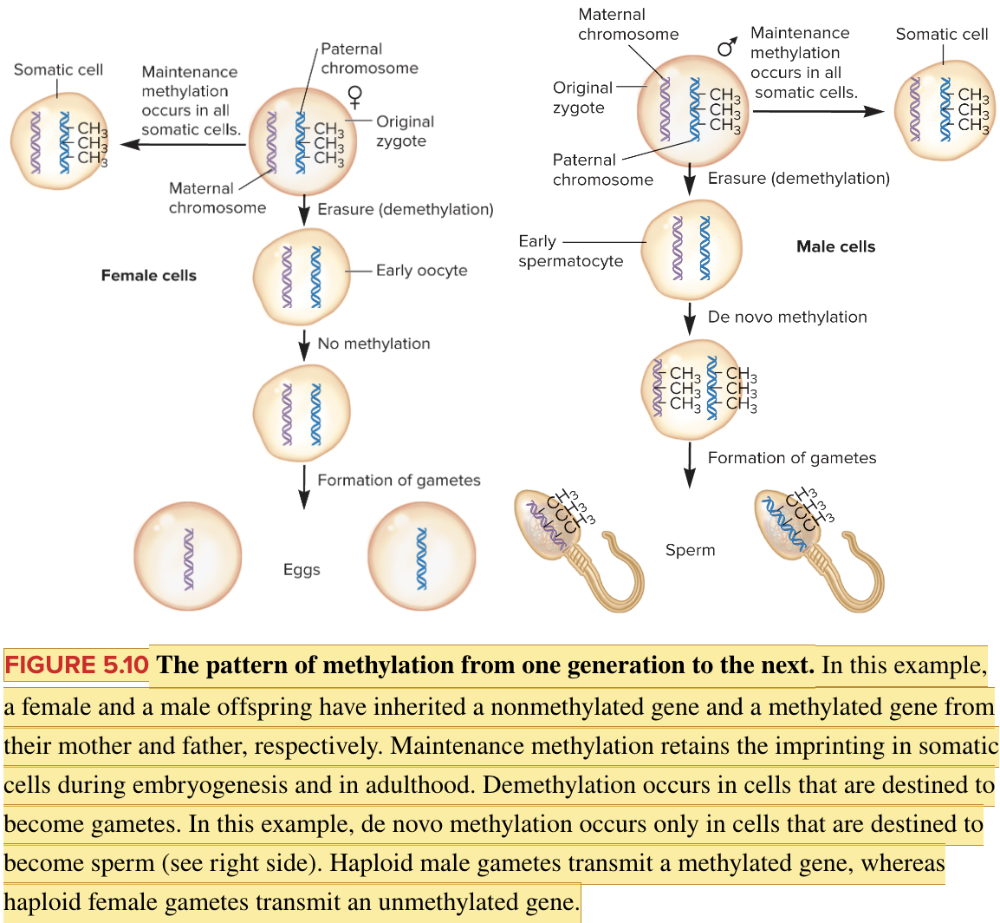 Maternal Reason: The DNA in the sperm has been imprinted by DNA methylation and will therefore be silenced. |
front 54 Select all that apply What information do you need to predict an offspring's phenotype when considering imprinted genes? Multiple select question. Which allele was inherited from which parent The genotypes of the offspring's siblings Which allele is on the Barr body of the female parent If the offspring expresses the allele that is inherited from the mother or the father | back 54 Which allele was inherited from which parent If the offspring expresses the allele that is inherited from the mother or the father |
front 55 Match each stage of genomic imprinting with the correct description. Establishment of the imprint Maintenance of the imprint Erasure and reestablishment of the imprint The imprint is removed in germline cells During gametogenesis either maternal or paternal allele is silenced and the other is expressed The imprint pattern is continued throughout development in somatic cells | back 55 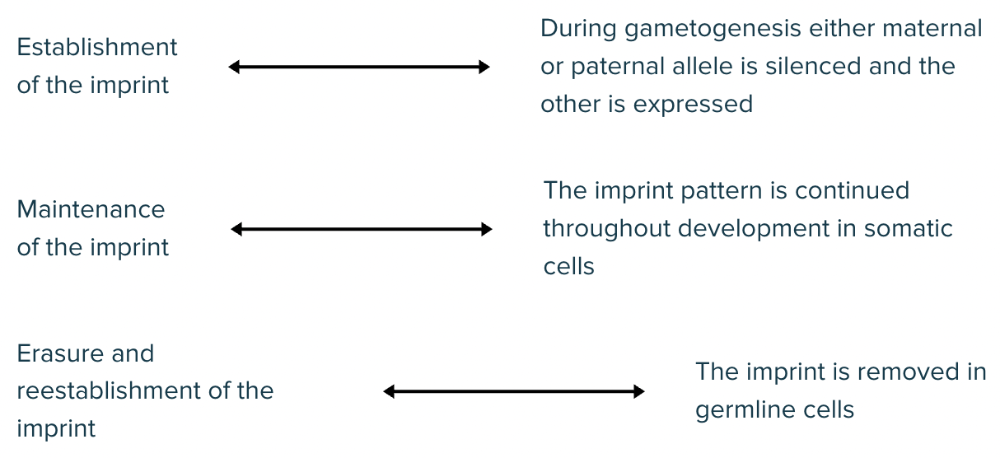
|
front 56 Select all that apply Select all of these that are human diseases caused by genomic imprinting. Multiple select question. Angelman syndrome Leber hereditary optic neuropathy Mitochondrial myopathy Prader-Willi syndrome | back 56 Angelman syndrome Prader-Willi syndrome |
front 57 In a somatic cell, the genomic imprint is _____. Multiple choice question. temporary permanent | back 57 permanent |
front 58 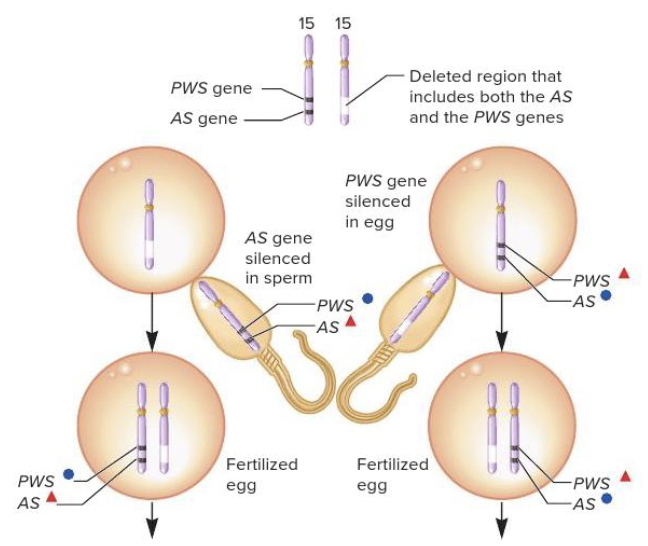 Based on the genomic imprints in the figure, the fertilized egg on the right will have which genetic disease? Multiple choice question. Angelman syndrome Prader-Willi syndrome | back 58 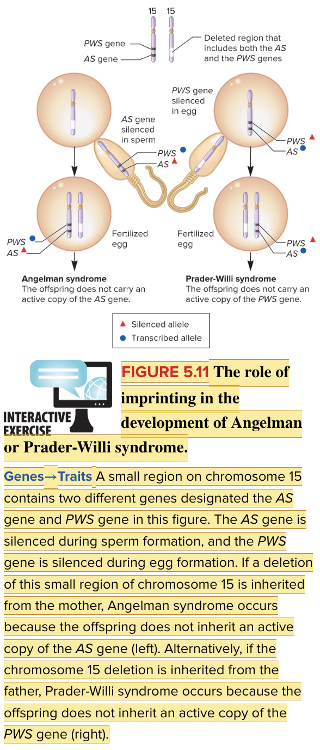 Prader-Willi syndrome Reason: If a chromosome 15 deletion is inherited from the father, Prader-Willi syndrome occurs because the offspring does not inherit an active copy of the PWS gene. |
front 59 The regulation of a genomic imprint involves ______ near the imprinted gene. Multiple choice question. an imprinting control region an origin of replication a promoter region a start codon | back 59 an imprinting control region |
front 60 For Angelman syndrome, which gene is lacking in expression? Multiple choice question. snoRNA genes UBE3A NDN SNRPN | back 60 UBE3A |
front 61 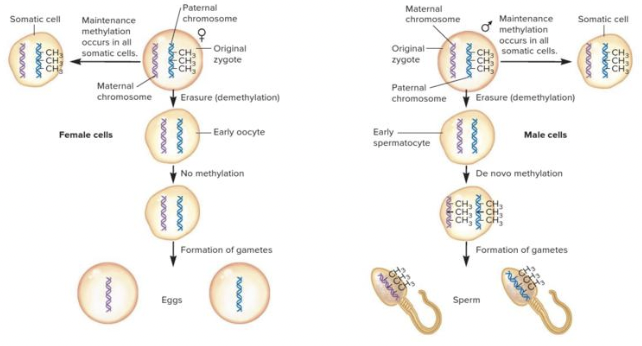 As seen in the figure, de novo methylation of a gene or chromosome occurs in ______. Multiple choice question. somatic cells gametes | back 61 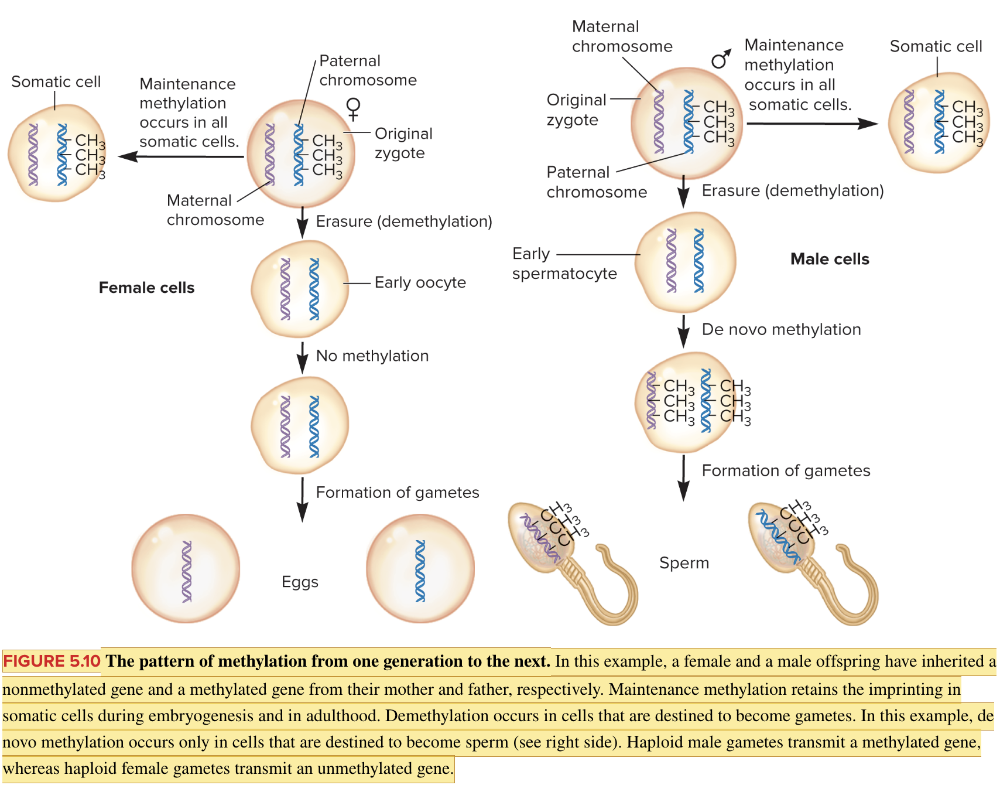 gametes |
front 62 Prader-Willi syndrome is associated with the imprinting of a gene encoding a splicing factor called ______. Multiple choice question. snoRNA SNRPN NDN UBE3A | back 62  SNRPN |
front 63 Select all that apply Which of these are examples of extranuclear genes? Multiple select question. Genes located on a chromosome in the nucleus Genes located on a chromosome in a mitochondrion Genes located on a chromosome in a chloroplast | back 63 Genes located on a chromosome in a mitochondrion Genes located on a chromosome in a chloroplast |
front 64 The human diseases Prader-Willi syndrome and Angelman syndrome are influenced by ______. Multiple choice question. maternal effect heteroplasmy genomic imprinting paternal leakage | back 64 genomic imprinting Reason: These diseases depend on inheriting a specific chromosomal deletion either from the father or mother. |
front 65 Fill in the blank question. An offspring receives genetic material located in the mitochondria. This is an example of ____ inheritance. | back 65 Blank 1: extranuclear, cytoplasmic, maternal, or nonmendelian |
front 66  Based on the genomic imprints in the figure, the fertilized egg on the left will have which genetic disease? Multiple choice question. Angelman syndrome Prader-Willi syndrome | back 66  Angelman syndrome Reason: If a chromosome 15 deletion is inherited from the mother, Angelman syndrome occurs because the offspring does not inherit an active copy of the AS gene. |
front 67 A mitochondrion or chloroplast ______ one nucleoid. Multiple choice question. may have more than one will only have one | back 67 may have more than one |
front 68  A deletion of a region of chromosome 15 that includes the gene ____ has been shown to cause Angelman syndrome. | back 68  UBE3A |
front 69 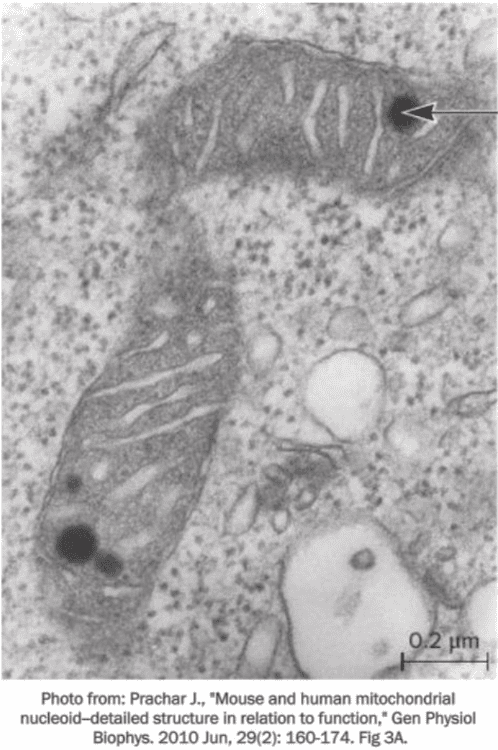 Fill in the blank question. In a mitochondrion, genetic material is located in a region called the ____, as indicated by the arrow in the image. | back 69 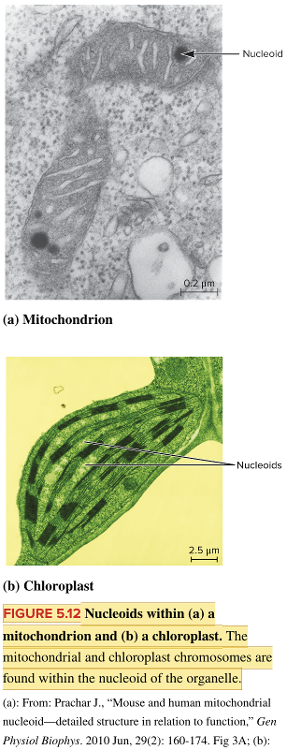 nucleoid |
front 70 Select all that apply Which chromosome 15 genes are lost in the deletion causing Prader-Willi syndrome? Multiple select question. SNRPN NDN UBE3A snoRNA genes | back 70 SNRPN NDN snoRNA genes |
front 71 Genes located on chromosomes are called nuclear genes because they are located in the cell | back 71 nucleus |
front 72 Select all that apply Select all of these that are true of mitochondrial genes. Multiple select question. Most mitochondrial proteins are encoded by nuclear genes. Human mtDNA carries relatively few genes. Mitochondrial DNA contains only coding regions. All mitochondrial proteins are encoded by mitochondrial genes. Mitochondrial DNA contains both coding and noncoding regions. | back 72 Most mitochondrial proteins are encoded by nuclear genes. Human mtDNA carries relatively few genes. Mitochondrial DNA contains both coding and noncoding regions. |
front 73 Select all that apply The genetic material inherited in an organelle, such as a mitochondrion or a chloroplast, exhibits _____ inheritance. Multiple select question. extranuclear mendelian cytoplasmic nuclear | back 73 extranuclear cytoplasmic |
front 74 True or false: An organelle such as a mitochondrion or a chloroplast will only have one nucleoid. | back 74 False Reason: Organelles often have more than one nucleoid. Chloroplasts found in Euglena may have 20-34 nucleoids. |
front 75 The genome of a chloroplast is _____ than the genome of a mitochondrion. Multiple choice question. the same size as larger smaller | back 75 larger |
front 76 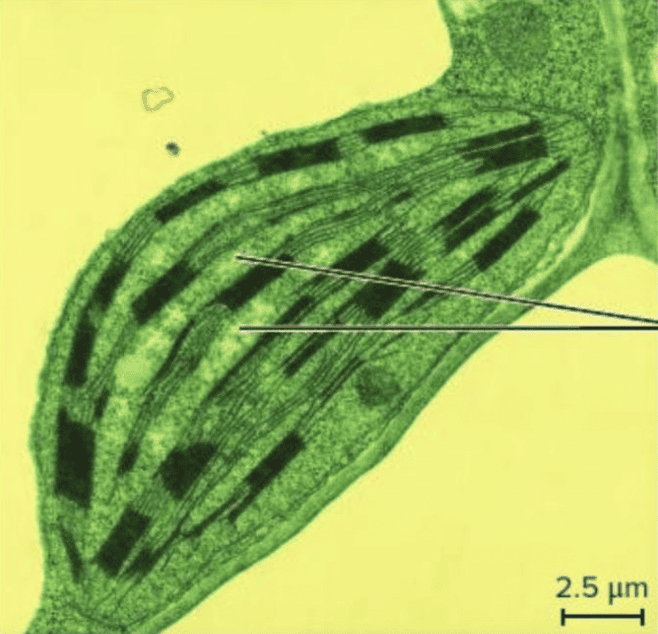 In chloroplasts, DNA is found in regions called ____, as indicated by the lines in the image. | back 76  nucleoid |
front 77 True or false: The genes located in mitochondria and chloroplasts do not exhibit Mendelian patterns of inheritance. | back 77 True Reason: Extranuclear genes are not segregated into gametes the same way the nuclear chromosomes are. As such, extranuclear genes do not exhibit Mendelian patterns of inheritance. |
front 78 Prader-Willi syndrome is associated with the imprinting of a gene encoding a splicing factor called ______. Multiple choice question. NDN snoRNA UBE3A SNRPN | back 78 SNRPN |
front 79 The type of inheritance that occurs because extranuclear genes are inherited through the cytoplasm of an egg is called | back 79 Blank 1: maternal, extranuclear, or cytoplasmicBlank 2: inheritance or inheritence |
front 80 The size of a human mitochondrial chromosome is ______ the size of a typical bacterial chromosome. Multiple choice question. more than 10% less than 1% equal to | back 80 less than 1% |
front 81 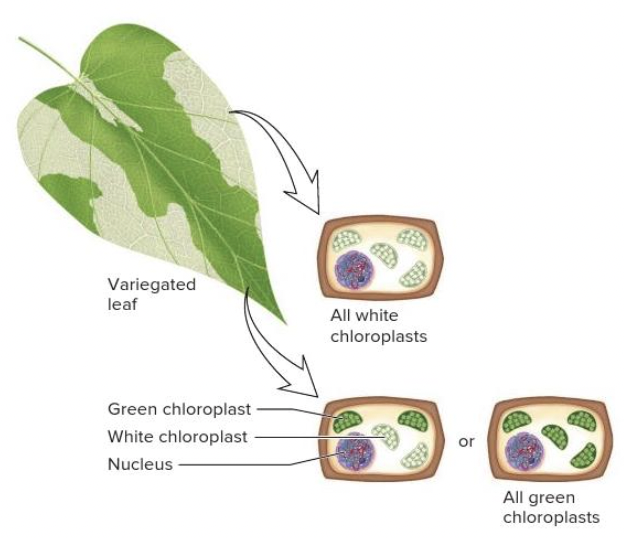 Variegation, a condition in which a leaf will have a mixture of normal and mutant chloroplasts (as shown), is a condition known as | back 81 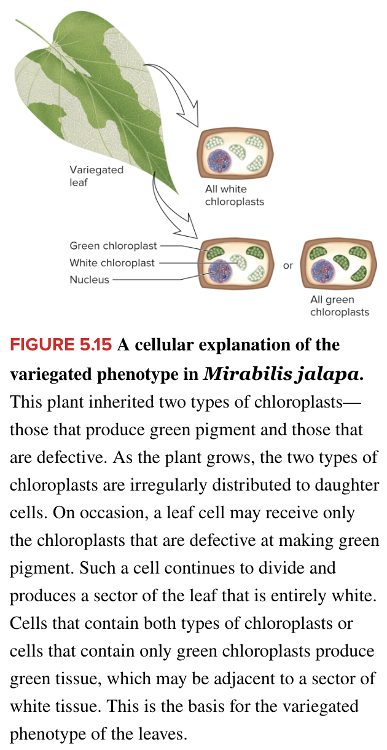 heteroplasmy |
front 82 Fill in the blank question. In ____ species, two kinds of gametes are made such that the distribution of cytoplasmic organelles is unequal in the two gamete types and organelles are often inherited from one parent. | back 82 heterogamous |
front 83 Select all that apply Select all of these that are true of chloroplast DNA. Multiple select question. The chloroplast genome is smaller than the mitochondrial genome. All chloroplast proteins are encoded by chloroplast genes. A typical chloroplast genome is about 100,000 - 200,000 bp in length. Many chloroplast proteins are encoded by nuclear genes. | back 83 A typical chloroplast genome is about 100,000 - 200,000 bp in length. Many chloroplast proteins are encoded by nuclear genes. |
front 84 In a species where maternal inheritance is the norm, the rare case in which mitochondria are provided by the sperm is a phenomenon called | back 84 paternal leakage |
front 85 Extranuclear genes are not segregated into gametes the same way the nuclear chromosomes are. As such, extranuclear genes ______ exhibit Mendelian patterns of inheritance. Multiple choice question. do do not | back 85 do not |
front 86 In chloroplasts, the gene for the photosynthetic enzyme RUBISCO can be mutated. Leaf cells with chloroplasts carrying this mutation will appear white due to the absence of photosynthesis. Leaves with a mixture of mutated and non-mutated chloroplasts show heteroplasmy, with green and white variegation. The RUBISCO gene is inherited from the mother. If the mother plant has green leaves and the father plant has white leaves, which types of leaves can be seen among the offspring? Multiple choice question. white, green, and variegated green white white and green but not variegated variegated | back 86 green |
front 87 Chloroplasts and mitochondria are inherited through the cytoplasm of the egg. This type of extranuclear inheritance is called ______. Multiple choice question. genomic imprinting heteroplasmy maternal inheritance maternal effect | back 87 maternal inheritance |
front 88 Select all that apply Select characteristics of mitochondrial DNA mutations. Multiple select question. Mitochondrial DNA mutations may occur in somatic cells and accumulate as a person ages. Mitochondrial DNA mutations are only inherited from the father. Mitochondrial DNA mutations are transmitted from a mother only to her male offspring. Mitochondrial DNA mutations are only found in gamete-forming cells, not in somatic cells. Mitochondrial DNA mutations are transmitted from a mother to all of her offspring. | back 88 Mitochondrial DNA mutations may occur in somatic cells and accumulate as a person ages. Mitochondrial DNA mutations are transmitted from a mother to all of her offspring. |
front 89 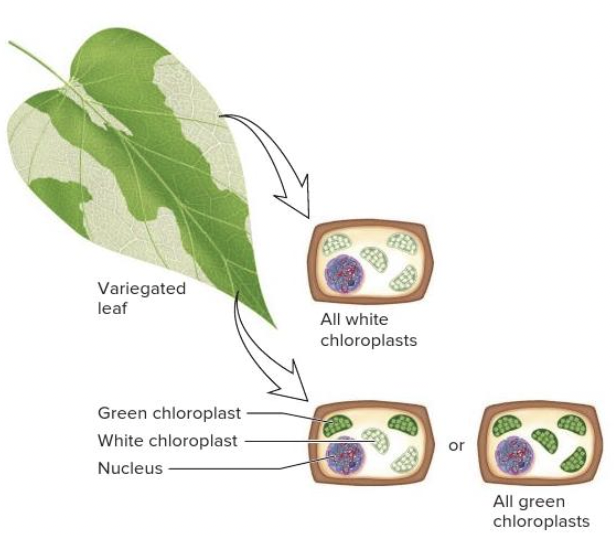 The variegation of the leaf shown in the image can be explained by ______. Multiple choice question. X-chromosome inactivation heteroplasmy paternal leakage Mendelian inheritance | back 89  heteroplasmy |
front 90 Human mitochondrial diseases, such as Leber hereditary optic neuropathy, are usually ______ inherited. Multiple choice question. maternally paternally | back 90 maternally |
front 91 A species that produces two kinds of gametes, with unequal distribution of cytoplasmic organelles, is said to be a ______ species. Multiple choice question. codominant incompletely penetrant heterogamous incompletely dominant | back 91 heterogamous |
front 92 Select all that apply Which human diseases are caused by mutations in mitochondrial DNA? Multiple select question. Angelman syndrome Neurogenic muscle weakness Prader-Willi syndrome Leber hereditary optic neuropathy | back 92 Neurogenic muscle weakness Leber hereditary optic neuropathy |
front 93 For a human offspring to express a paternal mitochondrial gene, the individual must have inherited a mitochondrion from the sperm. This is called ______. Multiple choice question. paternal leakage maternal leakage incomplete penetrance incomplete dominance | back 93 paternal leakage |
front 94 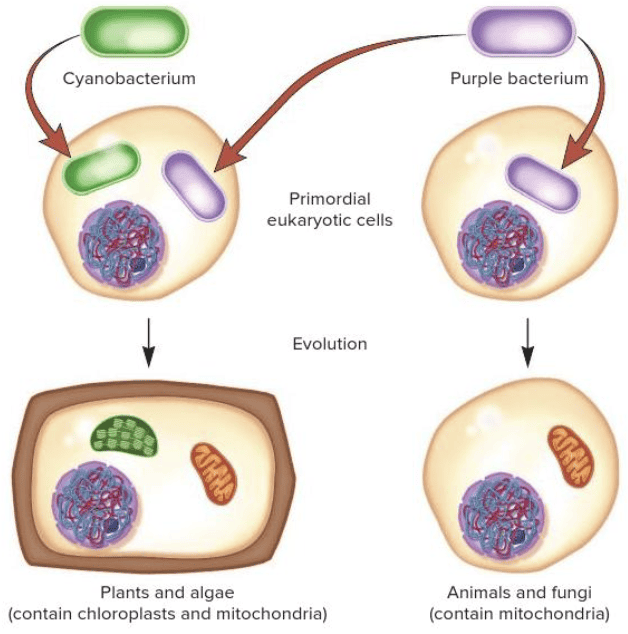 Fill in the blank question. The theory that explains the evolution of some eukaryotic organelles such as chloroplasts and mitochondria is called the ____ theory. | back 94 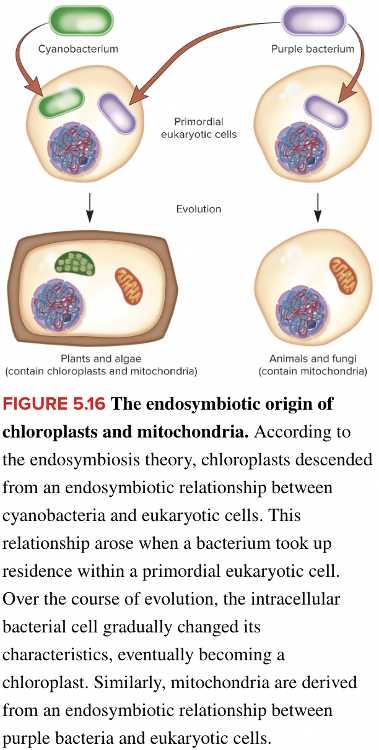 Blank 1: endosymbiosis or endosymbiotic |
front 95 In chloroplasts, the gene for the photosynthetic enzyme RUBISCO can be mutated. Leaf cells with chloroplasts carrying this mutation will appear white due to the absence of photosynthesis. Leaves with a mixture of mutated and non-mutated chloroplasts show heteroplasmy, with green and white variegation. The RUBISCO gene is inherited from the mother. If the mother plant is variegated and the father plant has fully green leaves, which types of leaves can be seen among the offspring? Multiple choice question. variegated only green and white but not variegated variegated and green but not white green, white, and variegated white only green only | back 95 green, white, and variegated |
front 96 Which inheritance pattern is typical of the transmission of mutations in mitochondrial DNA in humans? Multiple choice question. From the father to his daughters only From the mother to her sons only From the mother to her daughters only From the mother to all of her offspring From the father to all of his offspring From the father to his sons only | back 96 From the mother to all of her offspring |
front 97 Mitochondria may have evolved from an endosymbiotic relationship between ______. Multiple choice question. purple bacteria and eukaryotic cells yeast and purple bacteria algae and eukaryotic cells cyanobacteria and purple bacteria | back 97  purple bacteria and eukaryotic cells Reason: Both cyanobacteria and purple bacteria were involved in the endosymbiotic origin of chloroplasts and mitochondria. However, they do not exhibit an endosymbiotic relationship with each other. Mitochondria evolved from an endosymbiotic relationship between purple bacteria and eukaryotic cells. Chloroplasts evolved from an endosymbiotic relationship between cyanobacteria and eukaryotic cells. |
front 98 Select all that apply Which of these statements are true for human mitochondrial diseases? Multiple select question. They are usually chronic degenerative disorders They result from genomic imprinting They usually affect cells that require high levels of ATP They can usually be overcome by functional genes in the cell nucleus They often exhibit varying degrees of severity within families | back 98 They are usually chronic degenerative disorders They usually affect cells that require high levels of ATP They often exhibit varying degrees of severity within families |
front 99 The variation of gene expression that is not related to the variation of the DNA, is transmissible, and is reversible is described as | back 99 epigenetics |
front 100 Which human mitochondrial disease is caused by a mutation in a gene required for ATP synthesis? Multiple choice question. Mitochondrial myopathy Leber hereditary optic neuropathy Maternal myopathy and cardiomyopathy Neurogenic muscle weakness | back 100 Neurogenic muscle weakness |
front 101 Fill in the blank question. The endosymbiosis theory explains how eukaryotic cells developed the cellular organelles: ____ and ____. | back 101 mitochondria; chloroplasts |
front 102 True or false: Because epigenetics refers to heritable changes in gene expression, all epigenetic changes are examples of transgenerational epigenetic inheritance. | back 102 False Reason: Epigenetic changes may be permanent over an individual's life but are not permanent over multiple generations. |
front 103 Select all that apply Which of the following are examples of epigenetics? Multiple select question. DNA methylation Localization of histone variants DNA translocation Covalent histone modification Chromatin remodeling DNA mutation | back 103 DNA methylation Localization of histone variants Covalent histone modification Chromatin remodeling Reason: Epigenetics is the variation of gene expression that is not related to the variation of the DNA. |
front 104 Chloroplasts evolved from an endosymbiotic relationship between ______. Multiple choice question. purple bacteria and eukaryotic cells algae and eukaryotic cells cyanobacteria and eukaryotic cells cyanobacteria and purple bacteria yeast and purple bacteria | back 104  cyanobacteria and eukaryotic cells Reason: Both cyanobacteria and purple bacteria were involved in the endosymbiotic origin of chloroplasts and mitochondria. However, they do not exhibit an endosymbiotic relationship with each other. Mitochondria evolved from an endosymbiotic relationship between purple bacteria and eukaryotic cells. Chloroplasts evolved from an endosymbiotic relationship between cyanobacteria and eukaryotic cells. |
front 105 How do transcription factors contribute to epigenetic modification? Multiple choice question. Transcription factors recruit chromatin modifying proteins, such as DNA methyltransferase. Transcription factors allow mutated coding sequences to be transcribed. Transcription factors permanently bind to the DNA preventing the expression of specific gene sequences. Transcription factors bind to DNA inducing the removal of specific DNA sequences. | back 105 Transcription factors recruit chromatin modifying proteins, such as DNA methyltransferase. Reason: Transcription factors temporarily bind to DNA and typically increase the level of transcription. |
front 106 Select all that apply Which of the following are true regarding epigenetics? Multiple select question. Variations are reversible from one generation to the next Variations of gene expression result from mutations Epigenetic changes may be transmitted to offspring Variations of gene expression are unrelated to variations in the DNA base sequence Epigenetic changes are transmissible from cell to cell | back 106 Variations are reversible from one generation to the next Epigenetic changes may be transmitted to offspring Variations of gene expression are unrelated to variations in the DNA base sequence Epigenetic changes are transmissible from cell to cell |
front 107 Genes that are expressed but do not encode polypeptides produce ____-____ RNAs. | back 107 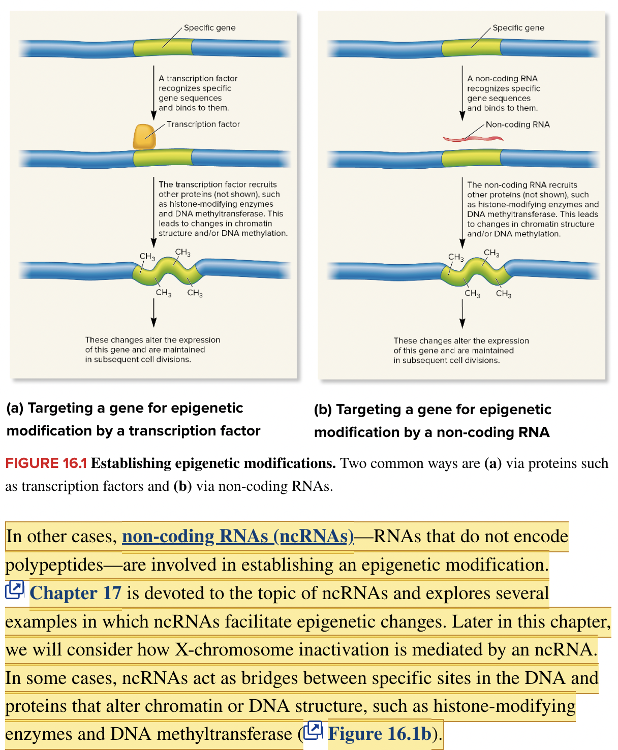 non-coding |
front 108 Select all that apply Which human diseases are caused by mutations in mitochondrial DNA? Multiple select question. Neurogenic muscle weakness Prader-Willi syndrome Leber hereditary optic neuropathy Angelman syndrome | back 108 Neurogenic muscle weakness Leber hereditary optic neuropathy |
front 109 If an epigenetic change is maintained only at a given site and does not affect the expression of a gene elsewhere in the nucleus, it is a ______ mechanism. Multiple choice question. trans-epigenetic cis-epigenetic | back 109 cis-epigenetic Reason: Trans-epigenetic mechanisms occur when a gene is activated that encodes a transcription factor that is capable of stimulating its own expression. The transcription factor may also turn on other genes. |
front 110 Which of the following is an example of epigenetic inheritance? Multiple choice question. A modified histone in a stem cell A chromatin modification that causes cancer in a lung cell Methylation of DNA that occurs in an oocyte Mutation of DNA in a sperm cell | back 110 Methylation of DNA that occurs in an oocyte Reason: Stem cells are somatic cells that will pass changes to other somatic cells but not to offspring. Reason: Changes that cause cancer in somatic cells are not transmitted to offspring. Reason: Changes in DNA sequence are genetic, rather than epigenetic, changes. |
front 111 Select all that apply Trans-epigenetic mechanisms are more commonly found in ______. Multiple select question. plants single-celled eukaryotes multicellular eukaryotes bacteria animals | back 111 single-celled eukaryotes bacteria Reason: Trans-epigenetic mechanisms are more commonly found in prokaryotes (bacteria and archaea) and single celled eukaryotes. |
front 112 DNA methylation, chromatin remodeling, covalent histone modification, and localization of histone variants are all examples of molecular mechanisms that are involved in | back 112 epigenetics |
front 113 Which of the following statements regarding X chromosome inactivation is true? Multiple choice question. X chromosome inactivation occurs during embryogenesis in both females and XY males. X chromosome inactivation occurs during embryogenesis in females. X chromosome inactivation occurs during embryogenesis in XY males. | back 113 X chromosome inactivation occurs during embryogenesis in females. |
front 114 True or false: Gene regulation may be related to epigenetic events such as the binding of transcription factors to certain genes, which induce permanent mutations in the DNA. | back 114 False Reason: Epigenetics due to changes in gene expression that do not involve changes in DNA sequence. Transcription factors recruit proteins that produce changes in gene expression but do not lead to mutations. |
front 115 Select all that apply Which statements are examples of how environmental factors may function as epigenetic regulators? Multiple select question. Phenotypic changes occur due to a spontaneous frameshift mutation that alters protein folding. Vernalization occurs when certain flowering plants are exposed to colder temperatures during the previous winter. Genomic imprinting during development leads to phenotypic changes. Body differences between queen bees and worker bees are caused by dietary differences. Exposure to tobacco smoke has been shown to alter DNA methylation and covalent modifications of specific genes in lung cells. | back 115 Vernalization occurs when certain flowering plants are exposed to colder temperatures during the previous winter. Body differences between queen bees and worker bees are caused by dietary differences. Exposure to tobacco smoke has been shown to alter DNA methylation and covalent modifications of specific genes in lung cells. |
front 116 An RNA that does not encode a polypeptide is a(n) ______ RNA. Multiple choice question. immature secondary non-coding structural | back 116 non-coding |
front 117 Select all that apply Which of the following are characteristics of euchromatin? Multiple select question. Localized along the periphery of the cell nucleus Inhibition of gene expression Highly compact structure High level of gene expression Occupies a central position in the nucleus | back 117 High level of gene expression Occupies a central position in the nucleus |
front 118  Which type of epigenetic mechanism is shown in the image? Multiple choice question. cis-epigenetic mechanism trans-epigenetic mechanism | back 118 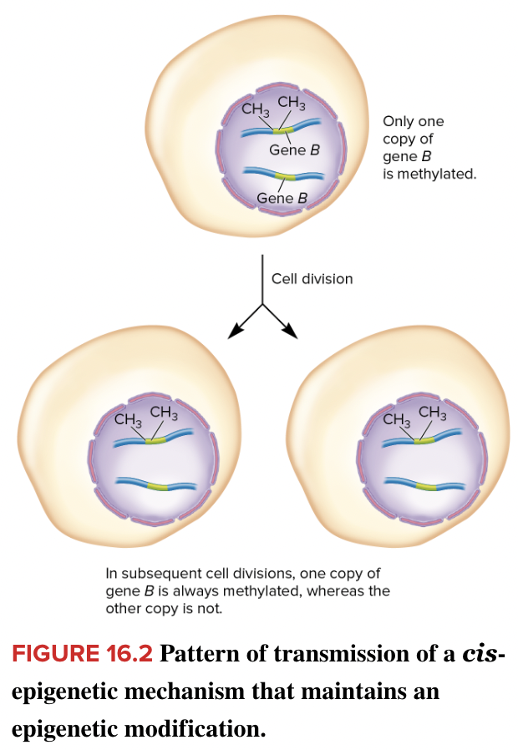 cis-epigenetic mechanism Reason: If an epigenetic change is maintained only at a given site and does not affect the expression of a gene elsewhere in the nucleus it is a cis-epigenetic mechanism. Reason: Trans-epigenetic mechanisms occur when a gene is activated that encodes a transcription factor that is capable of stimulating its own expression. The transcription factor may also turn on other genes. |
front 119 Select all that apply The role of heterochromatin in repressing transcription may be due to interference with which of the following factors? Multiple select question. General transcription factors Transcriptional activators Coactivators RNA polymerase II | back 119 General transcription factors Transcriptional activators Coactivators |
front 120 Activation of a gene that encodes a transcription factor capable of stimulating its own expression and/or the expression of other genes is an example of a ______ mechanism. Multiple choice question. cis-epigenetic trans-epigenetic | back 120 trans-epigenetic Reason: A cis-epigenetic change is maintained only at a given site and does not affect the expression of a gene elsewhere in the nucleus. |
front 121 Genomic imprinting and X-chromosome inactivation are examples of ______ mechanisms that occur during development. Multiple choice question. trans-epigenetic cis-epigenetic | back 121 cis-epigenetic Reason: Trans-epigenetic mechanisms occur when a gene is activated that encodes a transcription factor that is capable of stimulating its own expression. The transcription factor may also turn on other genes. |
front 122 How does heterochromatin prevent the movement of transposable elements (TEs)? Multiple choice question. Promoting histone acetylation in chromatin containing TEs Silencing genes needed for transposition Removing nucleosomes from regions containing TEs Preventing DNA replication of TEs | back 122 Silencing genes needed for transposition |
front 123 Select all that apply Which of the following are environmental factors that promote epigenetic changes? Multiple select question. Temperature Genomic imprinting X chromosome inactivation Diet Toxins | back 123 Temperature Diet Toxins Reason: This is an example of an epigenetic change rather than an environmental factor promoting the change. |
front 124 Characteristics of heterochromatin include which of the following? Multiple select question. Inhibition of gene expression Localized along the periphery of the cell nucleus Highly compact structure Occupies a central position in the nucleus High level of gene expression | back 124 Inhibition of gene expression Localized along the periphery of the cell nucleus Highly compact structure |
front 125 Select all that apply Identify ways that heterochromatin can inhibit transcription. Multiple select question. Preventing activators from binding to enhancer sequences Inhibiting the recruitment of general transcription factors or coactivators Creating nucleosome-free regions near promoters Increasing the level of DNA methylation in promoter regions | back 125 Preventing activators from binding to enhancer sequences Inhibiting the recruitment of general transcription factors or coactivators |
front 126 How does heterochromatin prevent the proliferation of viruses? Multiple choice question. Preventing the replication of DNA regions containing proviral DNA Enhancing the integration of the virus into the DNA of the host cell Inhibiting the expression of viral genes needed to produce new viruses Preventing the packaging of viral DNA into the viral protein coat | back 126 Inhibiting the expression of viral genes needed to produce new viruses |
front 127 Select all that apply Trans-epigenetic mechanisms are more commonly found in ______. Multiple select question. bacteria single-celled eukaryotes plants multicellular eukaryotes animals | back 127 bacteria (prokaryotes) single-celled eukaryotes Reason: Trans-epigenetic mechanisms are more commonly found in prokaryotes (bacteria and archaea) and single celled eukaryotes. |
front 128 Fill in the blank question. Regions of heterochromatin that vary among different cell types are called heterochromatin. | back 128  facultative |
front 129 When sites containing transposable elements (TEs) are converted to heterochromatin, the TEs are prevented from moving because the genes for transposition are ______. Multiple choice question. silenced over expressed mutated activated | back 129 silenced |
front 130 Regions of the chromatin containing numerous, short, tandemly repeated sequences that are located near the centromeres and at the telomeres of eukaryotic chromosomes are called ______ heterochromatin. Multiple choice question. euchromatic facultative constitutive activated | back 130 constitutive |
front 131 Fill in the blank question. Regions of a protein that catalyze the addition of posttranslational modifications (PTMs) are called ____ domains. | back 131 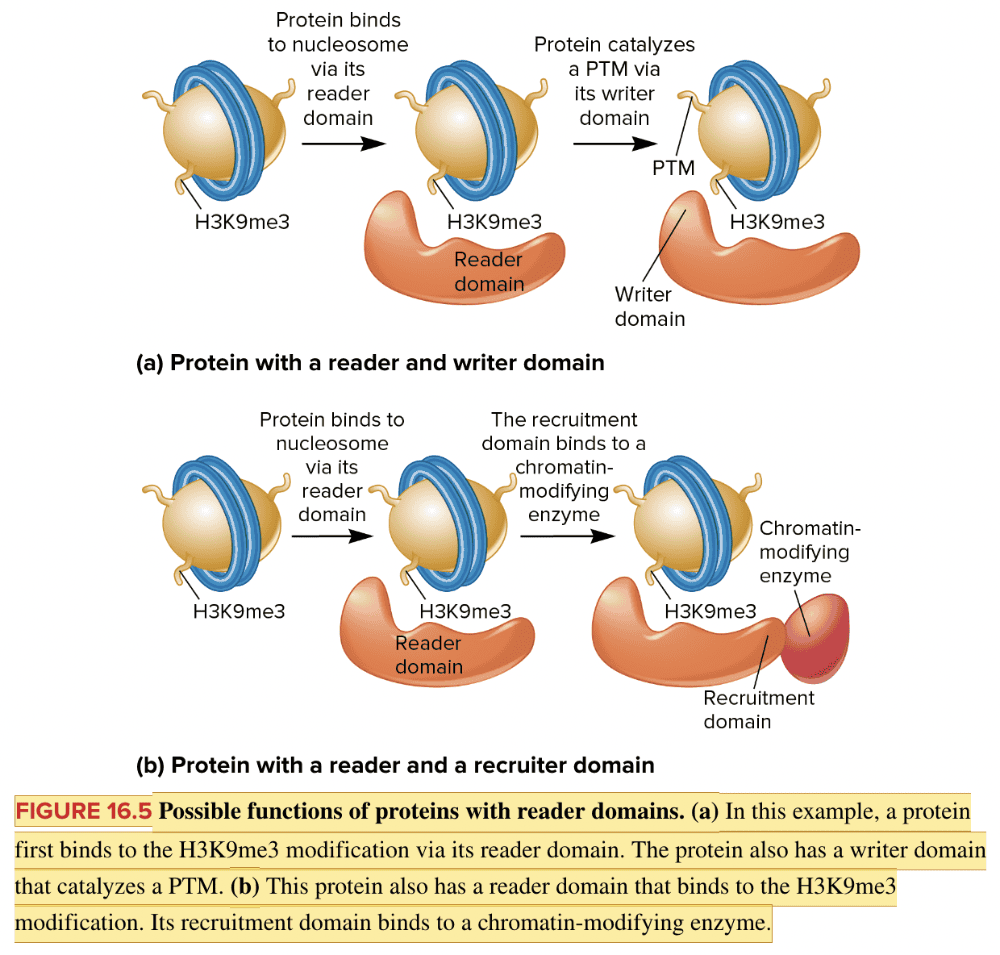 writer |
front 132 In the prevention of viral proliferation, what affect does heterochromatin have on a virus that has integrated into the host genome as proviral DNA? Multiple choice question. Prevents nucleosome formation near the promoters of proviral genes Prevents expression of proviral genes needed to produce new viruses Causes the excision of proviral DNA from the host genome Causes mutations in proviral DNA that result in the formation of faulty products | back 132 Prevents expression of proviral genes needed to produce new viruses |
front 133 Hypoacetylation of lysines in histone tails is a common modification found in ______. Multiple choice question. transcriptionally active regions facultative euchromatin constitutive heterochromatin constitutive euchromatin | back 133 constitutive heterochromatin |
front 134 Fill in the blank question. Regions of heterochromatin that are seen in the same location in all cell types are called ____ heterochromatin. | back 134 constitutive |
front 135 When bound to a nucleosome carrying a modified histone, a binding protein with a reader domain but no writer domain will ______. Multiple choice question. recruit chromatin-modifying enzymes reposition the surrounding nucleosomes catalyze a posttranslational modification methylate the associated DNA | back 135 recruit chromatin-modifying enzymes |
front 136 Heterochromatic regions found at multiple discrete sites located between the centromeres and telomeres, and which contain methylated DNA at CpG islands in regulatory regions, are called ______ heterochromatin. Multiple choice question. facultative euchromatic constitutive activated | back 136 facultative |
front 137 Select all that apply Which of the following are features associated with constitutive heterochromatin in yeast and animal cells? Multiple select question. Composed of many, short tandemly repeated sequences Located at telomeres and close to centromeres Significant number of nucleosome-free regions near gene promoters Trimethylation of a lysine at the ninth position in histone H3 DNA is highly methylated on cytosines DNA is composed of unique gene sequences | back 137 Composed of many, short tandemly repeated sequences Located at telomeres and close to centromeres Trimethylation of a lysine at the ninth position in histone H3 DNA is highly methylated on cytosines |
front 138 Match the type of chromatin structure with the stage of the cell cycle in which it would be observed. Interphase M phase M phase through interphase in two daughter cells Euchromatic regions condense into constitutive and facultative heterochromatin. Constitutive and facultative heterochromatin regions retain the same pattern observed in mother cell. Most chromosomal regions are composed of euchromatin. | back 138 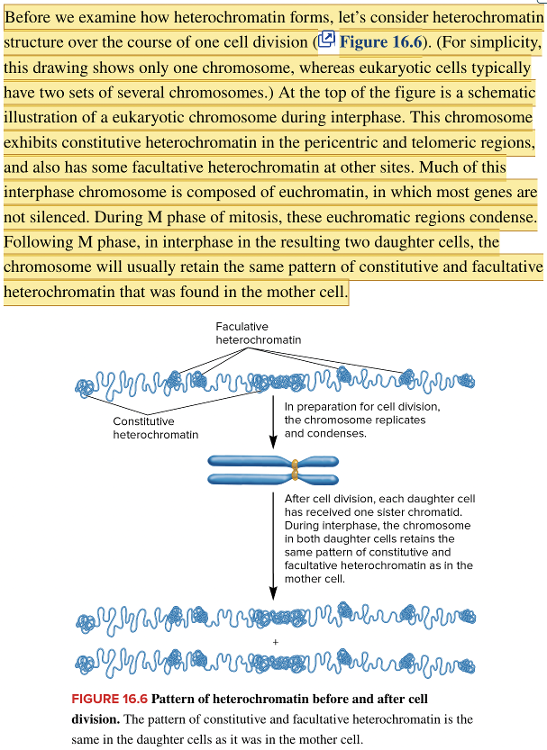
|
front 139 Regions of a protein that control the binding of particular proteins to nucleosomes with posttranslational modifications (PTMs) are called ______ domains. Multiple choice question. reader eraser sensor writer | back 139 reader |
front 140 Fill in the blank question. Any assemblage of nucleosomes that assumes a reproducible confirmation in three-dimensional space represents a(n) ____-____ structure. | back 140 higher-order |
front 141 Which modification is commonly found for lysines in histones associated with constitutive heterochromatin? Multiple choice question. hypomethylation hyperacetylation hypoacetylation | back 141 hypoacetylation |
front 142 Once bound to a nucleosome carrying a modified histone, a binding protein with both a reader and writer domain will ______. Multiple choice question. catalyze a posttranslational modification methylate the associated DNA recruit chromatin-modifying enzymes reposition the surrounding nucleosomes | back 142 catalyze a posttranslational modification |
front 143 Select all that apply Which of the following are features associated with facultative heterochromatin? Multiple select question. Trimethylation of histone H3 at the 9th or 27th lysine Located at telomeres and close to centromeres Methylation at CpG islands located in the regulatory regions of genes Composed of many, short tandemly repeated sequences In animals, contain LINE-type repeated sequences Located at multiple discrete sites between the centromere and telomeres | back 143 Trimethylation of histone H3 at the 9th or 27th lysine Methylation at CpG islands located in the regulatory regions of genes In animals, contain LINE-type repeated sequences Located at multiple discrete sites between the centromere and telomeres |
front 144 Select all that apply Which molecular processes will enhance the formation of higher-order structure in heterochromatin? Multiple select question. Formation of nucleosome-free regions Unwinding of DNA from histone octamers Posttranslational modifications of histones Binding of proteins to nucleosomes DNA methylation | back 144 Posttranslational modifications of histones Binding of proteins to nucleosomes DNA methylation |
front 145 Fill in the blank question. During cell division, from M phase in the mother cell to interphase in the resulting two daughter cells, a chromosome will usually retain the same pattern of ____ and ____ heterochromatin that was found in the mother cell. | back 145 Blank 1: constitutive Blank 2: facultative |
front 146 Select all that apply Higher-order heterochromatin structure is characterized by ______. Multiple select question. closer contacts between nucleosomes formation of loop domains establishing nucleosome-free regions hypomethylation of DNA and histones binding of heterochromatin to the nuclear lamina | back 146 formation of loop domains closer contacts between nucleosomes binding of heterochromatin to the nuclear lamina |
front 147 Which of the following represents a higher-order structure found in chromatin? Multiple choice question. 30-nm fiber Histone octamer Extended chain of nucleosomes Nucleosome DNA double helix | back 147 30-nm fiber |
front 148 When binding to chromatin, where would a dimer of heterochromatin protein 1 (HP1) be found? Multiple choice question. In the linker region between nucleosomes Attached to the DNA that is coiled around the histone octamer Spanning a coiled region containing four nucleosomes | back 148 In the linker region between nucleosomes |
front 149 Hypoacetylation of lysines in histone tails is a common modification found in ______. Multiple choice question. facultative euchromatin transcriptionally active regions constitutive euchromatin constitutive heterochromatin | back 149 constitutive heterochromatin |
front 150 When bound to a nucleosome carrying a modified histone, a binding protein with a reader domain but no writer domain will ______. Multiple choice question. reposition the surrounding nucleosomes recruit chromatin-modifying enzymes catalyze a posttranslational modification methylate the associated DNA | back 150 recruit chromatin-modifying enzymes |
front 151 Fill in the blank question. Formation of higher-order structures in heterochromatin depends upon posttranslational modifications of ____, binding of proteins to ____, and the process of DNA ____. | back 151 Blank 1: histones Blank 2: nucleosomes Blank 3: methylation |
front 152 Fill in the blank question. Heterochromatin protein 1 (HP1) causes changes in heterochromatin structure when HP1 dimers ____ chromatin-modifying enzymes and chromatin-remodeling complexes to the site of dimer binding. | back 152 recruit or attract |
front 153 Fill in the blank question. Characteristics of higher-order heterochromatin structure include closer contact between ____, formation of ____ domains and binding of heterochromatin to the nuclear ____. | back 153 Blank 1: nucleosomes Blank 2: loop Blank 3: lamina |
front 154 Condensin and CCCTC-binding factor (CTCF) are involved in forming ____ domains in heterochromatin. | back 154 loop |
front 155 A protein that forms a dimer in the linker region between nucleosomes carrying the modified histone H3K9me3 is ______. Multiple choice question. euchromatin protein 1 (EU1) heterochromatin protein 1 (HP1) histone H1 CCCTC-binding factor (CTCF) | back 155 heterochromatin protein 1 (HP1) |
front 156 A fibrous layer of proteins lining the inner nuclear membrane of eukaryotic cells is called the ______. Multiple choice question. nuclear envelope nuclear lamina nucleolus nuclear matrix | back 156 nuclear lamina |
front 157 Select all that apply Once heterochromatin protein 1 (HP1) forms a dimer in the linker region between nucleosomes, which types of proteins does it recruit to the site? Multiple select question. Chromatin-remodeling complexes RNA polymerases Histones with posttranslational modifications Chromatin-modifying enzymes | back 157 Chromatin-remodeling complexes Chromatin-modifying enzymes |
front 158 Chromosomal regions that are in close contact with the nuclear lamina are called lamina-associated | back 158 domains |
front 159 Select all that apply Identify proteins involved in forming loop domains in heterochromatin. Multiple select question. CCCTC-binding protein condensin heterochromatin protein 1 (HP1) histone H1 H3K9me3 | back 159 CCCTC-binding protein condensin |
front 160 Select all that apply Select the roles that lamina-associated domains (LADs) are thought to fulfill. Multiple select question. Organize interphase chromosomes into chromosome territories Inhibit gene expression Enhance gene expression Promote higher levels of chromatin condensation | back 160 Organize interphase chromosomes into chromosome territories Inhibit gene expression |
front 161 In most eukaryotic cells, the inner nuclear membrane of the cell is lined by a fibrous layer of proteins called the nuclear | back 161 lamina |
front 162 Select all that apply Which characteristics are observed for lamina-associated domains (LADs)? Multiple select question. Found in both constitutive and facultative heterochromatin Contain a large number of nucleosome-free regions Contain about 2500 nucleosomes Show high levels of histone modification Found only in constitutive heterochromatin | back 162 Found in both constitutive and facultative heterochromatin Contain about 2500 nucleosomes Show high levels of histone modification |
front 163 Hypoacetylation of lysines in histone tails is a common modification found in ______. Multiple choice question. constitutive heterochromatin constitutive euchromatin transcriptionally active regions facultative euchromatin | back 163 constitutive heterochromatin |
front 164 Which components can initiate heterochromatin formation at a nucleation site? Multiple select question. sequence-specific DNA binding proteins chromatin-modifying proteins non-coding RNA H3K9me3 Reason: Trimethylation of H3 results from the nucleation process. CCCTC-binding factors (CTCFs) | back 164 sequence-specific DNA binding proteins non-coding RNA Reason: Trimethylation of H3 results from the nucleation process. |
front 165 Lamina-associated domains (LADs) are regions of ______ found in close contact with the nuclear lamina in eukaryotic cells. Multiple choice question. chromatin-modifying enzymes chromosomes replicating enzymes chromatin-remodeling complexes | back 165 chromosomes |
front 166 The enzyme that is recruited by heterochromatin protein 1 (HP1) during the spreading phase of heterochromatin formation is histone ______. Multiple choice question. acetyltransferase kinase methyltransferase deacetylase | back 166 methyltransferase |
front 167 Fill in the blank question. Organizing chromosomes into chromosome territories and inhibiting gene expression are two roles of the ____-____ domains found on eukaryotic chromosomes. | back 167 Blank 1: lamina Blank 2: associated |
front 168 Fill in the blank question. A typical eukaryotic chromosome contains a few dozen lamina-associated domains (LADs) found in both ____ and ____ heterochromatin. | back 168 Blank 1: constitutive Blank 2: facultative |
front 169 Select all that apply Which of the following can act as barriers to prevent spreading of heterochromatin into adjacent euchromatin regions? Multiple select question. a region that is fully covered by nucleosomes a nucleosome-free region an enhancer sequence a region containing many antisilencing proteins | back 169 a nucleosome-free region a region containing many antisilencing proteins |
front 170 Select all that apply In heterochromatin formation, which of the following are responsible for recruiting histone deacetylases and histone methyltransferases to a nucleation site? Multiple select question. CCCTC-binding factors (CTCFs) Non-coding RNAs H3K9me3 heterochromatin protein 1 (HP1) Sequence-specific DNA-binding proteins | back 170 Non-coding RNAs Sequence-specific DNA-binding proteins |
front 171 During development, the silencing of genes that should not be expressed in a specific cell type occurs by the formation of cell-specific ______ heterochromatin regions. Multiple choice question. constitutive facultative nucleated | back 171 facultative |
front 172 Select all that apply During the formation of heterochromatin, which proteins are involved in the self-propagating mechanism responsible for the spreading phase? Multiple select question. heterochromatin protein 1 (HP1) CCCTC-binding factor (CTCF) histone methyltransferase histone acetyltransferase | back 172 heterochromatin protein 1 (HP1) histone methyltransferase |
front 173 A heterochromatic region with fully methylated DNA and extensively modified histone H3 prior to DNA replication, will have ______ (the) levels of these modifications in the daughter chromatids produced after DNA replication. Multiple choice question. the same 1/2 1/4 | back 173 1/2 |
front 174 Fill in the blank question. One type of barrier to the spreading of heterochromatin is a region with histone-modifying enzymes that catalyze ____ or ____ of histones to promote the formation of euchromatin and inhibit the formation of heterochromatin. | back 174 Blank 1: demethylation Blank 2: acetylation |
front 175 Fill in the blank question. Heterochromatic regions that are cell-specific and silence genes that should not be expressed in a certain cell type are called ____ heterochromatin. | back 175 facultative |
front 176 Fill in the blank question. DNA methylation and histone trimethylation are mechanisms that quickly reestablish ____ structure after DNA replication. | back 176 heterochromatin |
front 177 Select all that apply Before replication, a heterochromatic region has sites with fully methylated DNA and nucleosomes with H3K9me3 histone modifications. After DNA replication, what would be the constituents of each daughter chromatid? Multiple select question. fully methylated DNA hemimetylated DNA half of the H3 histones with H3K9me3 modifications all of the H3 histones with H3K9me3 modifications | back 177 hemimetylated DNA half of the H3 histones with H3K9me3 modifications |
front 178 ICF syndrome and Roberts syndrome are inherited diseases associated with abnormalities in ______. Multiple choice question. heterochromatin formation formation of lamina-associated domains (LADs) nucleosome formation euchromatin activation | back 178 heterochromatin formation |
front 179 Select all that apply Abnormalities in heterochromatin formation have resulted from loss-of function mutations affecting the activity of which enzymes? Multiple select question. histone deacetylase acetyltransferase DNA methyltransferase histone methyltransferase | back 179 acetyltransferase DNA methyltransferase |
front 180 Select all that apply The Igf2 gene will be expressed if ______. Multiple select question. the ICR and DMR are methylated CTC factors bind to the ICR and DMR the Igf2 gene can be stimulated by the enhancer the H19 gene is methylated | back 180 the ICR and DMR are methylated the Igf2 gene can be stimulated by the enhancer |
front 181 Select all that apply Select mechanisms responsible for reestablishing heterochromatin structure after DNA replication. Multiple select question. Presence of higher-order heterochromatin structure Trimethylation of histones DNA methylation Deacetylation of histones Formation of nucleosome-free regions | back 181 Presence of higher-order heterochromatin structure Trimethylation of histones DNA methylation |
front 182 Select all that apply Idenfity the diseases that have been associated with abnormalities in heterochromatin formation. Multiple select question. ICF syndrome Cancer Roberts syndrome Cri-du-chat syndrome | back 182 ICF syndrome Roberts syndrome |
front 183 The methylation of a previously unmethylated site is called ______. Multiple choice question. de novo methylation de novo mutation maintenance methylation induced mutation | back 183 de novo methylation Reason: The methylation of hemimethylated sites is called maintenance methylation. |
front 184 Select all that apply Which two areas of the X chromosome play a key role in X inactivation? Multiple select question. Tsix Xist Igf2 H19 | back 184 Tsix Xist |
front 185 Fill in the blank question. Two inherited human diseases related to abnormalities in heterochromatin formation are ICF syndrome due to a loss-of-function mutation in a DNA ____ enzyme related to constitutive heterochromatin formation, and Roberts syndrome due to a loss-of-function mutation in a(n) ____ enzyme involved in establishing sister-chromatid cohesion. | back 185 Blank 1: methyltransferase Blank 2: acetyltransferase |
front 186 What accounts for the molecular mechanism of imprinting of the Igf2 gene? Multiple choice question. Spontaneous mutation in the Igf2 gene Differential methylation during oogenesis and spermatogenesis Spontaneous mutation in the H19 gene Differential methylation during fetal development | back 186 Differential methylation during oogenesis and spermatogenesis Reason: The molecular mechanism of Igf2 imprinting is due to different patterns of methylation during spermatogenesis and oogenesis. |
front 187 What happens when pluripotency factors stimulate the expression of Tsix? Multiple choice question. CTCFs are expressed. H19 is inhibited. Igf2 is expressed. Xist is inhibited. | back 187 Xist is inhibited. |
front 188 The methylation of hemimethylated sites is called ______. Multiple choice question. an induced mutation a de novo mutation maintenance methylation de novo methylation | back 188 maintenance methylation Reason: The methylation of a previously unmethylated site is called de novo methylation. |
front 189 Select all that apply Which of the following statements regarding X inactivation are true? Multiple select question. Expression of Tsix from both X chromosomes inhibits the expression of Xist In very early embryos, both X chromosomes are active Pluripotency factors inhibit the expression of Tsix When the Xist gene is inactive the X chromosome is active | back 189 Expression of Tsix from both X chromosomes inhibits the expression of Xist In very early embryos, both X chromosomes are active When the Xist gene is inactive the X chromosome is active |
front 190 X-chromosome inactivation is an epigenetic event that occurs during ______. Multiple choice question. spermatogenesis the postnatal period embryogenesis oogenesis | back 190 embryogenesis |
front 191 Click and drag on elements in order List the events of X-chromosome inactivation in the correct sequence. putting the first event at the top. All pluripotency factors and CTCFs are shifted to one of the two X chromosomes The Tsix gene of the chromosome without pluripotency factors and CTCFs is inhibited; the inactivated chromosome is coated with Xist RNA X chromosomes pair briefly beginning at the Tsix gene Xist RNA recruits proteins that epigenetically modify specific sites within the Xi | back 191 1. X chromosomes pair briedly beginning at the Tsix
gene |
front 192 True or false: The epigenetic modifications that lead to silencing of an X chromosome are lost and reestablished following each cell division. | back 192 False Reason: The epigenetic modifications in Xi are maintained in subsequent cell divisions. |
front 193 Select all that apply Identify changes that can occur during embryonic development that will lead to a pattern of differential gene expression unique to a specific cell type. Multiple select question. Loss of DNA containing genes not required for a specific cell type Genetic mutations that disable genes whose products are not needed in a specific cell type Epigenetic changes that enable specific genes to be transcribed Epigenetic changes that cause specific genes to be permanently repressed | back 193 Epigenetic changes that enable specific genes to be transcribed Epigenetic changes that cause specific genes to be permanently repressed |
front 194 Select all that apply Which two complexes are the key regulators of epigenetic changes that occur during development? Multiple select question. Trithorax group Imprinting control region Polycomb group X-inactivation center | back 194 Trithorax group Polycomb group |
front 195 The inactivation of an X chromosome is a(n) ______ event. Multiple choice question. mutational environmentally-controlled epigenetic temperature-dependent | back 195 epigenetic |
front 196 Fill in the blank question. The PcG complex attaches three methyl groups to lysine at position 27 in histone H3 in a process known as ____. This mark represses gene expression. | back 196 trimethylation |
front 197 The first step of X inactivation occurs when X chromosomes pair briefly beginning at the ______. Multiple choice question. Tsix gene macroH2A Xist gene CTCFs Xi | back 197 Tsix gene Reason: Pairing occurs at the Tsix gene and pluripotency factors and CTCFs that were previously bound to both chromosomes shift entirely to one chromosome.The chromosome to which they shift becomes active. The other X chromosome does not express Tsix, which permits expression of Xist. |
front 198 Select all that apply What are the three ways that PRC1 may inhibit transcription? Multiple select question. Covalent modification of H2A by attraction of ubiquitin molecules Direct interaction with transcription factors Nucleosomes in the target gene may form knot-like structures Direct methylation of the nucleotides in the target gene | back 198 Covalent modification of H2A by attraction of ubiquitin molecules Direct interaction with transcription factors Nucleosomes in the target gene may form knot-like structures |
front 199 Select all that apply Which changes occur following the binding of Xist RNAs to the Xi? Multiple select question. Recruitment of DNA methyltransferases to Xi Histone variant macroH2A is incorporated into the nucleosomes along the Xi Recruitment of protein complexes to Xi Activation of Tsix and subsequent inhibition of Xist Covalent modification of specific sites in histone tails | back 199 Recruitment of DNA methyltransferases to Xi Histone variant macroH2A is incorporated into the nucleosomes along the Xi Recruitment of protein complexes to Xi Covalent modification of specific sites in histone tails |
front 200 Fill in the blank question. During embryonic development, many genes within a specific cell type will undergo epigenetic changes that enable them to be ____ or cause them to be permanently ____. | back 200 Blank 1: transcribed or expressed Blank 2: repressed or silenced |
front 201 Epigenetic changes due to the actions of PcG complexes ______. Multiple choice question. are maintained in subsequent cell divisions are transient and short lived have no effect on gene expression | back 201 are maintained in subsequent cell divisions |
front 202 Trithorax group (TrxG) complexes epigenetically influence development by ______ various genes. Multiple choice question. inhibiting activating | back 202 activating Reason: PcG complexes cause gene repression. |
front 203 The TrxG complex attaches three methyl groups to lysine at position 4 in histone H3 in a process known as trimethylation. This mark ______ gene expression. Multiple choice question. activates inhibits | back 203 activates |
front 204 The red1 gene in maize produces dark red corn kernels. The red1' allele is paramutagenic and results in light pink kernels. A plant with a red1 allele that has undergone paramutation would have ______ kernels. Multiple choice question. dark red light pink | back 204 light pink |
front 205 Click and drag on elements in order List the steps of how a gene may be targeted for silencing by PcG complexes in the correct sequence, putting the first step on top. Transcription is inhibited by preventing binding of RNA polymerase and PRC1 may be recruited to inhibit transcription in various ways PRE-binding protein binds to PRE and recruits PRC2 PRC2 catalyzes trimethylation of lysine 27 in histone H3 | back 205 1. Prebinding proteins to pre and recruits prc2 |
front 206 In maize, the red1 + allele can change the expression level of red1 allele. The red1 allele is the ______ allele. Multiple choice question. paramutable paramutagenic | back 206 paramutable Reason: The paramutagenic allele is the allele that can change the expression of another allele. In this case the paramutagenic allele is the red1+ allele. |
front 207 In maize, the dominant allele B-I encodes a transcription factor that regulates genes involved in producing purple color of corn stalks and husks. The allele can undergo a mutation to B' which changes the stalk and husk color from purple to green. In a cross between B-I B-I and B' B' homozygotes, the F1 heterozygotes all have green stalks and husks. This shows that the B' allele is ____ and the B-I allele is ____. | back 207 Blank 1: paramutagenic Blank 2: paramutable |
front 208 True or false: Epigenetic changes due to PcG complexes are transient and only present during development. | back 208 False Reason: Epigenetic changes due to the actions of PcG complexes are maintained in subsequent cell divisions. |
front 209 Select all that apply Which of the following factors determine the effects of paramutation? Multiple select question. The number of paramutagenic alleles in the genome The rate of paramutation of the paramutagenic allele The stability of the paramutagenic allele The effectiveness of DNA repair systems | back 209 The rate of paramutation of the paramutagenic allele The stability of the paramutagenic allele |
front 210 Polycomb group (PcG) complexes epigenetically influence development by ______ various genes. Multiple choice question. activating inhibiting | back 210 inhibiting |
front 211 The mop1 gene, which is required for paramutation in Arabidopsis, encodes ______ that produces siRNA for gene silencing. Multiple choice question. a DNA-dependent RNA polymerase an exonuclease an RNA-dependent RNA polymerase a DNA methyltransferase | back 211 an RNA-dependent RNA polymerase |
front 212 The bI gene in maize regulates pigment synthesis. The B-I allele is dominant and produces purple husks, but the B' allele is paramutagenic and results in green husks. A B-I B' heterozygous plant would be expected to have ______ husks. Multiple choice question. green purple | back 212  green |
front 213 In maize, the red1 + allele can change the expression level of red1 allele. The red1 + allele is the ____ allele. | back 213 paramutagenic |
front 214 In mice, the Agouti gene regulates ______. Multiple choice question. the deposition of yellow pigment in hair the sex of mice the size of mice the transcription of cell cycling proteins | back 214 the deposition of yellow pigment in hair |
front 215 In maize, the dominant allele B-I codes for a transcription factor that regulates genes involved in producing purple color in corn stalks and husks. The allele can undergo a mutation to B' which changes the stalk and husk color from purple to green. In a cross between B-I B-I and B' B' homozygotes, the F1 heterozygotes all have green stalks and husks. When these F1 individuals are crossed to B-I B-I homozygotes, all of the F2 offspring have green stalks and husks. The results for the F2 generation demonstrate a ______ paramutation of the ______ allele. Multiple choice question. secondary; B-I allele. secondary; B' allele. primary; B-I allele. primary; B' allele. | back 215 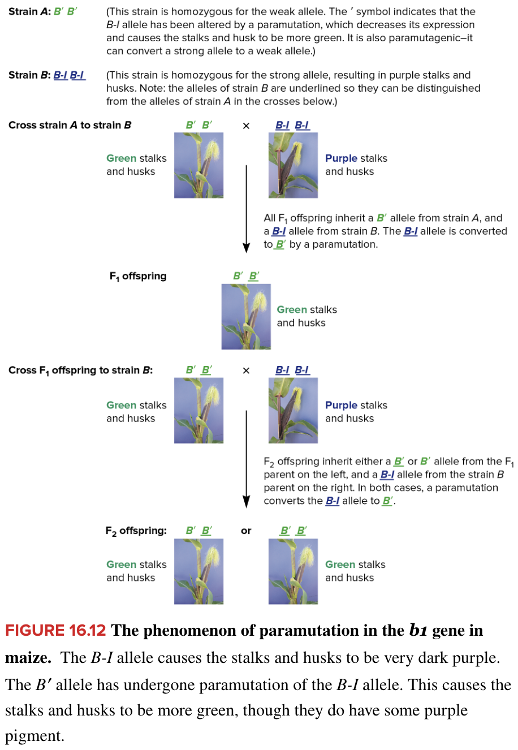 secondary; B-I allele. |
front 216 Mice that are homozygous for the loss-of-function mutation in the Agouti gene will have ______ fur. Multiple choice question. yellow black brown white | back 216 black Reason: A yellow band in each hair shaft is due to a dominant rather than a recessive allele. |
front 217 When a paramutation is passed down for several generations and continues to affect the expression of other alleles in each generation, this is known as ______. Multiple choice question. permanent paramutation stable paramutation secondary paramutation frequent paramutation | back 217 secondary paramutation Reason: There is a more specific term for this type (stable paramutation) of paramutation. |
front 218 Multiple tandem repeat sequences near paramutagenic alleles may be involved in paramutation by ______. Multiple choice question. binding many transcription factors to increase expression causing gene duplication to increase expression producing siRNAs to reduce expression causing frameshifts to alter the protein | back 218 producing siRNAs to reduce expression |
front 219 When there is a gain-of-function mutation in the mouse Agouti gene, the gene is ______ resulting in ______. Multiple choice question. not expressed; white fur over-expressed; black fur expressed normally; wild-type brown fur with black tips over-expressed; yellow fur | back 219 over-expressed; yellow fur |
front 220 Adult, wild-type mice with genotype AA for the Agouti gene will have what color coat hairs? Multiple choice question. All black Yellow with brown tips All yellow All brown Brown with black tips | back 220 Brown with black tips Reason: The yellow pigment is sandwiched between layers of black pigment, giving a brown color along most of the hair shaft. No yellow pigment is synthesized near the tip of the hair. Reason: No yellow pigment is synthesized near the tips of the hairs, so the tips will be black. |
front 221 What accounts for the wide array of coat colors seen in mice carrying the A vy allele of the Agouti gene? Multiple choice question. Mutation of the Agouti gene that regulates coat color Trimethylation of the Agouti gene that regulates coat color Epigenetic modification of the promoter in the transposable element responsible for over expressing the Agouti gene Variable duplication of the Agouti gene so that increasing copy number leads to lighter coloration | back 221 Epigenetic modification of the promoter in the transposable element responsible for over expressing the Agouti gene |
front 222 Mice that are homozygous for the loss-of-function mutation in the Agouti gene will have black fur because ______. Multiple choice question. melanin is not produced the mice are albino pheomelanin is not produced | back 222 pheomelanin is not produced |
front 223 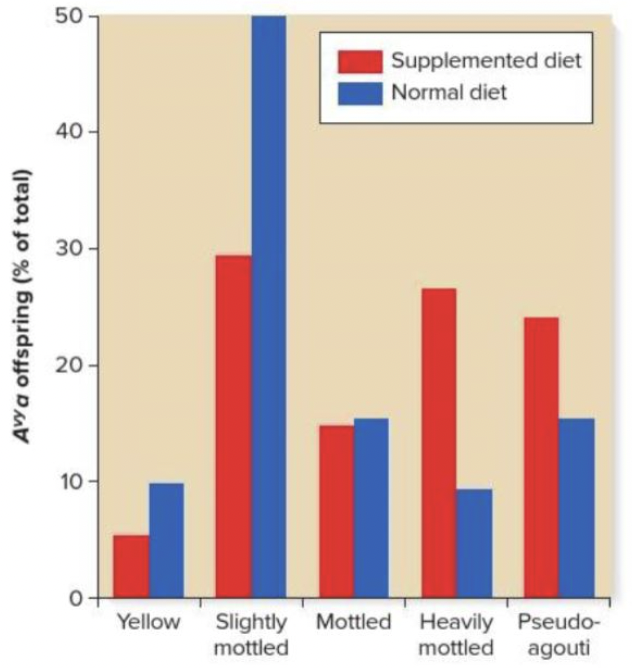 What can be deduced about the environmental effect of diet on expression of the Agouti gene? Multiple choice question. Offspring of females fed a supplemental diet exhibited the same color coat as the parents. Offspring of females fed a supplemental diet had darker coats. Offspring of females fed a supplemental diet had lighter coats. | back 223 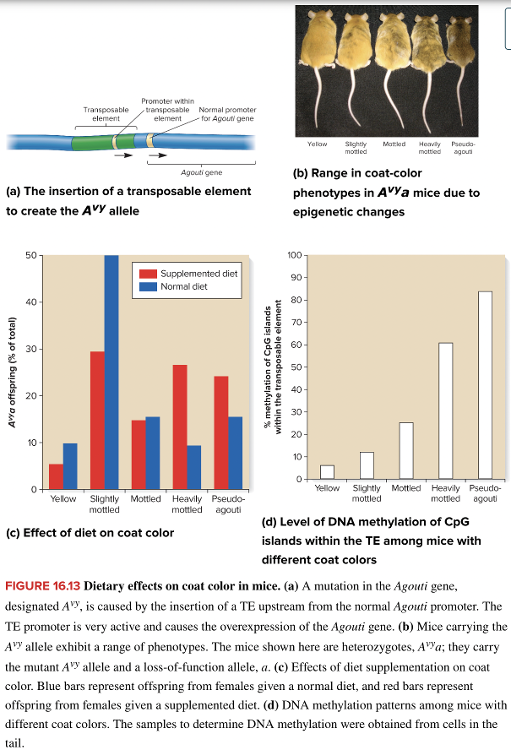 Offspring of females fed a supplemental diet had darker coats. |
front 224 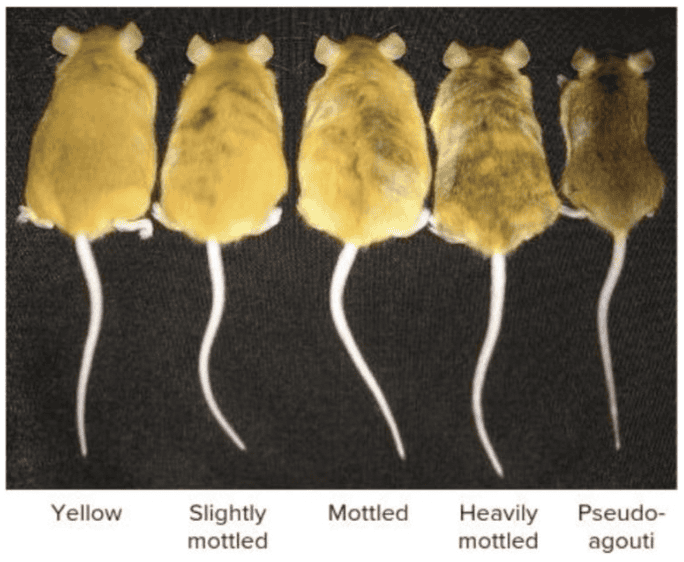 What accounts for the variation in coat color in the mice shown? Multiple choice question. Mutation within the coding region of the Agouti gene Degree of methylation at CpG islands in the TE upstream from the Agouti gene Degree of methylation within the Agouti gene Mutation in the promoter of the Agouti gene | back 224  Degree of methylation at CpG islands in the TE upstream from the Agouti gene Reason: All of the pictured mice carry the A vy allele for the Agouti gene Reason: Methylation of a region in the promoter, upstream of the gene, affects the level of coat color expression. |
front 225 Mice with a gain-of-function mutation in the Agouti gene will have ______ fur. Multiple choice question. black brown white yellow | back 225 yellow |
front 226 Female honeybee larvae fed royal jelly during early development, and nectar or pollen later in development will become ______ bees. Multiple choice question. worker queen | back 226 worker Reason: Queen bees need to be consistently fed on royal jelly. |
front 227 Mice with the A vy allele exhibit ______. Multiple choice question. completely white fur completely black fur a wide array of coat colors | back 227 a wide array of coat colors Reason: The transposable element associated with this allele appears to be sensitive to epigenetic modifications. |
front 228  What conclusion can be made regarding the environmental effect of diet on the expression of the Agouti gene? Multiple choice question. The supplemental diet influenced the expression of the Agouti gene. The supplemental diet had no effect on the expression of the Agouti gene. | back 228  The supplemental diet influenced the expression of the Agouti gene. |
front 229 The annual flowering plant Arabidopsis thaliana has two different types. Only the ______ require vernalization for flowering. Multiple choice question. summer-annuals, which grow from spring to fall, winter annuals, which grow from fall to spring, | back 229 winter annuals, which grow from fall to spring, |
front 230 Which transcription factors activate several genes that lead to flower development in the winter-annual type of Arabidopsis? Multiple choice question. VIN3 and COLDAIR FT and FLC FLC and SOC1 VRN1 and VRN2 FT and SOC1 | back 230 FLC and SOC1 Reason: Transcription of SOC1 is activated by another gene. |
front 231 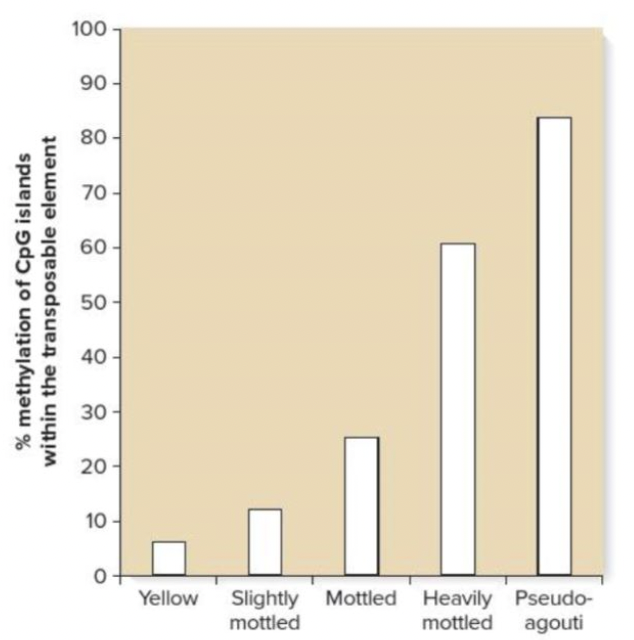 What can be deduced about the effect of DNA methylation levels on expression of the Agouti gene? Multiple choice question. Greater levels of methylation had no affect on coat color. Greater levels of methylation resulted in lighter coat color. Greater levels of methylation resulted in darker coat color. | back 231  Greater levels of methylation resulted in darker coat color. |
front 232 Female honeybee larvae fed and bathed in royal jelly during development will become ______ bees. Multiple choice question. queen worker | back 232 queen |
front 233 What accounts for the wide array of coat colors seen in mice carrying the A vy allele of the Agouti gene? Multiple choice question. Epigenetic modification of the promoter in the transposable element responsible for over expressing the Agouti gene Mutation of the Agouti gene that regulates coat color Variable duplication of the Agouti gene so that increasing copy number leads to lighter coloration Trimethylation of the Agouti gene that regulates coat color | back 233 Epigenetic modification of the promoter in the transposable element responsible for over expressing the Agouti gene |
front 234 Click and drag on elements in order Order the steps in the regulation of FLC expression by exposure to cold temperatures, putting the first step at the top. VIN3/PRC2 complex binds to COLDAIR RNA as it is transcribed from within an FLC intron VIN3 protein and PRC2 form a complex VIN3/PRC2 complex methylates histone H3 within the FLC gene Prolonged cold induces expression of VIN3 and COLDAIR FLC gene expression is repressed | back 234 1. Prolonged cold induces expression of VIN3 and COLD AIR 2. VIN3 protein and PRC2 form a complex 3. VIN3/PRC2 complex binds to COLDAIR RNA as it is transcribed from within an FLC intron 4. VIN3/PRC2 complex methylates histone H3 within the FLC gene 5. FLC gene expression is repressed |
front 235 Select all that apply After vernalization, plants ______. Multiple select question. have acquired epigenetic modifications in their genome. immediately begin flowering. have the ability to flower when conditions are favorable. have acquired mutations in their genome. | back 235 have acquired epigenetic modifications in their genome. have the ability to flower when conditions are favorable. |
front 236 The protein ______ which represses genes required for flowering in the winter-annual type of Arabidopsis, is inhibited by prolonged exposure to cold. Multiple choice question. FT COLDAIR FLC VIN3 | back 236 FLC |
front 237 Which one best represents the current model of how cold temperatures cause repression of the FLC gene? Multiple choice question. Cold temperatures inhibit the expression of a transcription factor that represses FLC Cold temperatures induce expression of a non-coding RNA, COLDAIR, which recruits a chromatin modifying complex to FLC The transcription factor FT is unable to bind to the FLC promoter in cold temperature Cold temperatures cause an overall decrease in plant gene expression | back 237 Cold temperatures induce expression of a non-coding RNA, COLDAIR, which recruits a chromatin modifying complex to FLC |